Functional equivalence of an evolutionarily conserved RNA binding module
- PMID: 26292216
- PMCID: PMC4591824
- DOI: 10.1074/jbc.M115.673012
Functional equivalence of an evolutionarily conserved RNA binding module
Abstract
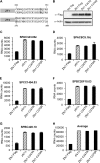
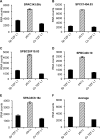
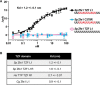
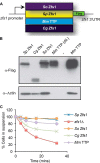
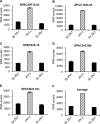
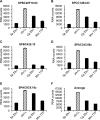
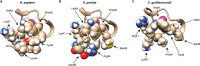
Members of the tristetraprolin (TTP) family of proteins participate in the regulation of mRNA turnover after initially binding to AU-rich elements in target mRNAs. Related proteins from most groups of eukaryotes contain a conserved tandem zinc finger (TZF) domain consisting of two closely spaced, similar CCCH zinc fingers that form the primary RNA binding domain. There is considerable sequence variation within the TZF domains from different family members within a single organism and from different organisms, raising questions about sequence-specific effects on RNA binding and decay promotion. We hypothesized that TZF domains from evolutionarily distant species are functionally interchangeable. The single family member expressed in the fission yeast Schizosaccharomyces pombe, Zfs1, promotes the turnover of several dozen transcripts, some of which are involved in cell-cell interactions. Using knockin techniques, we replaced the TZF domain of S. pombe Zfs1 with the equivalent domains from human TTP and the single family member proteins expressed in the silkworm Bombyx mori, the pathogenic yeast Candida guilliermondii, and the plant Chromolaena odorata. We found that the TZF domains from these widely disparate species could completely substitute for the native S. pombe TZF domain, as determined by measurement of target transcript levels and the flocculation phenotype characteristic of Zfs1 deletion. Recombinant TZF domain peptides from several of these species bound to an AU-rich RNA oligonucleotide with comparably high affinity. We conclude that the TZF domains from TTP family members in these evolutionarily widely divergent species are functionally interchangeable in mRNA binding and decay.
Keywords: RNA binding protein; RNA turnover; mRNA decay; yeast genetics; zinc finger.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

