Clonal Diversification and Changes in Lipid Traits and Colony Morphology in Mycobacterium abscessus Clinical Isolates
- PMID: 26292297
- PMCID: PMC4609687
- DOI: 10.1128/JCM.02015-15
Clonal Diversification and Changes in Lipid Traits and Colony Morphology in Mycobacterium abscessus Clinical Isolates
Abstract
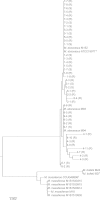
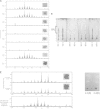
The smooth-to-rough colony morphology shift in Mycobacterium abscessus has been implicated in loss of glycopeptidolipid (GPL), increased pathogenicity, and clinical decline in cystic fibrosis (CF) patients. However, the evolutionary phenotypic and genetic changes remain obscure. Serial isolates from nine non-CF patients with persistent M. abscessus infection were characterized by colony morphology, lipid profile via thin-layer chromatography and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), sequencing of eight genes in the GPL locus, and expression level of fadD23, a key gene involved in the biosynthesis of complex lipids. All 50 isolates were typed as M. abscessus subspecies abscessus and were clonally related within each patient. Rough isolates, all lacking GPL, predominated at later disease stages, some showing variation within rough morphology. While most (77%) rough isolates harbored detrimental mutations in mps1 and mps2, 13% displayed previously unreported mutations in mmpL4a and mmpS4, the latter yielding a putative GPL precursor. Two isolates showed no deleterious mutations in any of the eight genes sequenced. Mixed populations harboring different GPL locus mutations were detected in 5 patients, demonstrating clonal diversification, which was likely overlooked by conventional acid-fast bacillus (AFB) culture methods. Our work highlights applications of MALDI-TOF MS beyond identification, focusing on mycobacterial lipids relevant in virulence and adaptation. Later isolates displayed accumulation of triacylglycerol and reduced expression of fadD23, sometimes preceding rough colony onset. Our results indicate that clonal diversification and a shift in lipid metabolism, including the loss of GPL, occur during chronic lung infection with M. abscessus. GPL loss alone may not account for all traits associated with rough morphology.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


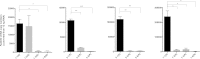
References
-
- Adekambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol 56:133–143. doi:10.1099/ijs.0.63969-0. - DOI - PubMed
-
- Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. doi:10.1128/JCM.01688-08. - DOI - PMC - PubMed
-
- Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi:10.1016/S0140-6736(13)60632-7. - DOI - PMC - PubMed
-
- Tettelin H, Davidson RM, Agrawal S, Aitken ML, Shallom S, Hasan NA, Strong M, de Moura VC, De Groote MA, Duarte RS, Hine E, Parankush S, Su Q, Daugherty SC, Fraser CM, Brown-Elliott BA, Wallace RJ Jr, Holland SM, Sampaio EP, Olivier KN, Jackson M, Zelazny AM. 2014. High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks Emerg Infect Dis 20:364–371. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

