Defining the three cell lineages of the human blastocyst by single-cell RNA-seq
- PMID: 26293300
- PMCID: PMC4582176
- DOI: 10.1242/dev.123547
Defining the three cell lineages of the human blastocyst by single-cell RNA-seq
Erratum in
-
Defining the three cell lineages of the human blastocyst by single-cell RNA-seq.Development. 2015 Oct 15;142(20):3613. doi: 10.1242/dev.131235. Development. 2015. PMID: 26487783 Free PMC article.
Abstract
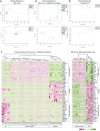
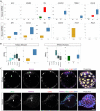
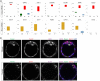
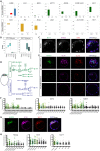
Here, we provide fundamental insights into early human development by single-cell RNA-sequencing of human and mouse preimplantation embryos. We elucidate conserved transcriptional programs along with those that are human specific. Importantly, we validate our RNA-sequencing findings at the protein level, which further reveals differences in human and mouse embryo gene expression. For example, we identify several genes exclusively expressed in the human pluripotent epiblast, including the transcription factor KLF17. Key components of the TGF-β signalling pathway, including NODAL, GDF3, TGFBR1/ALK5, LEFTY1, SMAD2, SMAD4 and TDGF1, are also enriched in the human epiblast. Intriguingly, inhibition of TGF-β signalling abrogates NANOG expression in human epiblast cells, consistent with a requirement for this pathway in pluripotency. Although the key trophectoderm factors Id2, Elf5 and Eomes are exclusively localized to this lineage in the mouse, the human orthologues are either absent or expressed in alternative lineages. Importantly, we also identify genes with conserved expression dynamics, including Foxa2/FOXA2, which we show is restricted to the primitive endoderm in both human and mouse embryos. Comparison of the human epiblast to existing embryonic stem cells (hESCs) reveals conservation of pluripotency but also additional pathways more enriched in hESCs. Our analysis highlights significant differences in human preimplantation development compared with mouse and provides a molecular blueprint to understand human embryogenesis and its relationship to stem cells.
Keywords: Embryonic stem cells; Epiblast; Human; Mouse; RNA-sequencing; Trophectoderm.
© 2015. Published by The Company of Biologists Ltd.
Figures



References
-
- Aanes H., Winata C. L., Lin C. H., Chen J. P., Srinivasan K. G., Lee S. G. P., Lim A. Y. M., Hajan H. S., Collas P., Bourque G. et al. (2011). Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 21, 1328-1338. 10.1101/gr.116012.110 - DOI - PMC - PubMed
-
- Albano R. M., Groome N. and Smith J. C. (1993). Activins are expressed in preimplantation mouse embryos and in ES and EC cells and are regulated on their differentiation. Development 117, 711-723. - PubMed
-
- Ang S. L., Wierda A., Wong D., Stevens K. A., Cascio S., Rossant J. and Zaret K. S. (1993). The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development 119, 1301-1315. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

