Single-cell ATAC-seq: strength in numbers
- PMID: 26294014
- PMCID: PMC4546161
- DOI: 10.1186/s13059-015-0737-7
Single-cell ATAC-seq: strength in numbers
Abstract
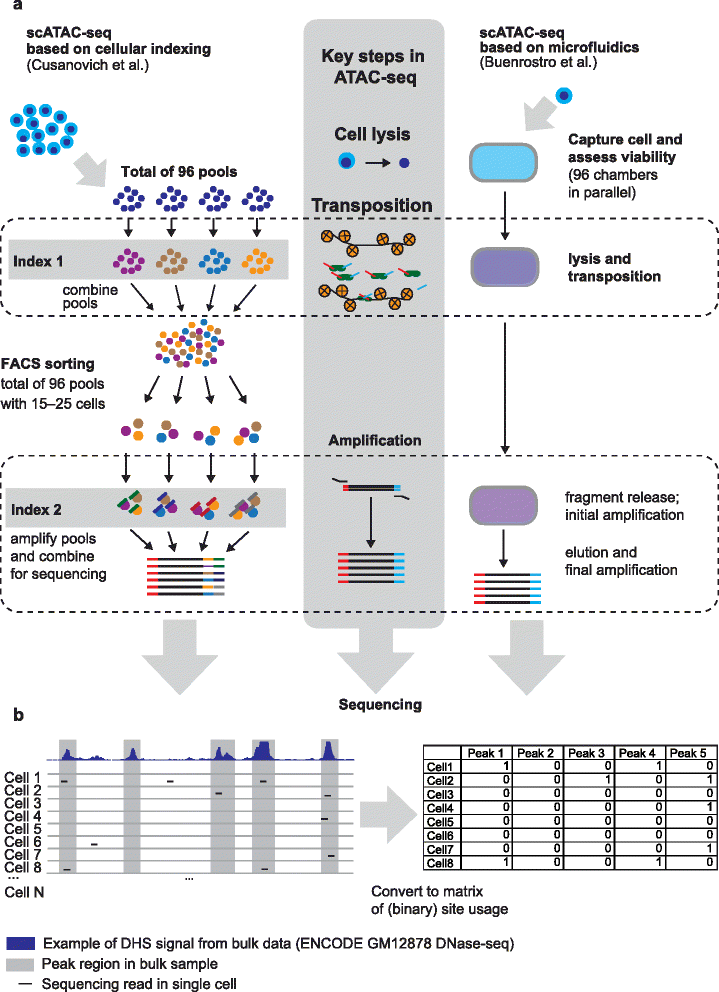
Single-cell ATAC-seq detects open chromatin in individual cells. Currently data are sparse, but combining information from many single cells can identify determinants of cell-to-cell chromatin variation.
Figures
Comment on
-
Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing.Science. 2015 May 22;348(6237):910-4. doi: 10.1126/science.aab1601. Epub 2015 May 7. Science. 2015. PMID: 25953818 Free PMC article.
-
Single-cell chromatin accessibility reveals principles of regulatory variation.Nature. 2015 Jul 23;523(7561):486-90. doi: 10.1038/nature14590. Epub 2015 Jun 17. Nature. 2015. PMID: 26083756 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

