Double Variational Binding--(SMILES) Conformational Analysis by Docking Mechanisms for Anti-HIV Pyrimidine Ligands
- PMID: 26295229
- PMCID: PMC4581313
- DOI: 10.3390/ijms160819553
Double Variational Binding--(SMILES) Conformational Analysis by Docking Mechanisms for Anti-HIV Pyrimidine Ligands
Abstract
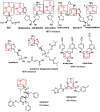
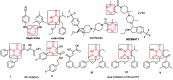
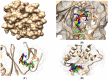
Variational quantitative binding-conformational analysis for a series of anti-HIV pyrimidine-based ligands is advanced at the individual molecular level. This was achieved by employing ligand-receptor docking algorithms for each molecule in the 1,3-disubstituted uracil derivative series that was studied. Such computational algorithms were employed for analyzing both genuine molecular cases and their simplified molecular input line entry system (SMILES) transformations, which were created via the controlled breaking of chemical bonds, so as to generate the longest SMILES molecular chain (LoSMoC) and Branching SMILES (BraS) conformations. The study identified the most active anti-HIV molecules, and analyzed their special and relevant bonding fragments (chemical alerts), and the recorded energetic and geometric docking results (i.e., binding and affinity energies, and the surface area and volume of bonding, respectively). Clear computational evidence was also produced concerning the ligand-receptor pocket binding efficacies of the LoSMoc and BraS conformation types, thus confirming their earlier presence (as suggested by variational quantitative structure-activity relationship, variational-QSAR) as active intermediates for the molecule-to-cell transduction process.
Keywords: 1,3-disubstituted uracil derivative; SMILES; anti-HIV; chemical binding affinity; interacting amino acid; ligand-receptor docking.
Figures





Similar articles
-
Determining chemical reactivity driving biological activity from SMILES transformations: the bonding mechanism of anti-HIV pyrimidines.Molecules. 2013 Jul 30;18(8):9061-116. doi: 10.3390/molecules18089061. Molecules. 2013. PMID: 23903183 Free PMC article.
-
Molecular docking, QSAR and ADMET based mining of natural compounds against prime targets of HIV.J Biomol Struct Dyn. 2019 Jan;37(1):131-146. doi: 10.1080/07391102.2017.1420489. Epub 2018 Jan 7. J Biomol Struct Dyn. 2019. PMID: 29268664
-
Computer-aided molecular design of highly potent HIV-1 RT inhibitors: 3D QSAR and molecular docking studies of efavirenz derivatives.SAR QSAR Environ Res. 2006 Aug;17(4):353-70. doi: 10.1080/10629360600884520. SAR QSAR Environ Res. 2006. PMID: 16920659
-
Rational design of HIV-1 entry inhibitors.Methods Mol Biol. 2013;993:185-204. doi: 10.1007/978-1-62703-342-8_13. Methods Mol Biol. 2013. PMID: 23568472 Review.
-
Latest trends in structure based drug design with protein targets.Adv Protein Chem Struct Biol. 2020;121:1-23. doi: 10.1016/bs.apcsb.2019.11.008. Epub 2019 Dec 18. Adv Protein Chem Struct Biol. 2020. PMID: 32312418 Review.
Cited by
-
Classification and Design of HIV-1 Integrase Inhibitors Based on Machine Learning.Comput Math Methods Med. 2021 Apr 1;2021:5559338. doi: 10.1155/2021/5559338. eCollection 2021. Comput Math Methods Med. 2021. PMID: 33868450 Free PMC article.
-
Structure-Based Design and in Silico Screening of Virtual Combinatorial Library of Benzamides Inhibiting 2-trans Enoyl-Acyl Carrier Protein Reductase of Mycobacterium tuberculosis with Favorable Predicted Pharmacokinetic Profiles.Int J Mol Sci. 2019 Sep 24;20(19):4730. doi: 10.3390/ijms20194730. Int J Mol Sci. 2019. PMID: 31554227 Free PMC article.
-
Multi-Layer Identification of Highly-Potent ABCA1 Up-Regulators Targeting LXRβ Using Multiple QSAR Modeling, Structural Similarity Analysis, and Molecular Docking.Molecules. 2016 Nov 29;21(12):1639. doi: 10.3390/molecules21121639. Molecules. 2016. PMID: 27916850 Free PMC article.
-
Structural Investigation for Optimization of Anthranilic Acid Derivatives as Partial FXR Agonists by in Silico Approaches.Int J Mol Sci. 2016 Apr 8;17(4):536. doi: 10.3390/ijms17040536. Int J Mol Sci. 2016. PMID: 27070594 Free PMC article.
-
Uracil derivatives as HIV-1 capsid protein inhibitors: design, in silico, in vitro and cytotoxicity studies.RSC Adv. 2022 Jun 13;12(27):17466-17480. doi: 10.1039/d2ra02450k. eCollection 2022 Jun 7. RSC Adv. 2022. PMID: 35765450 Free PMC article.
References
-
- Halford B. Aiming for HIV’s weak spot. Chem. Eng. News. 2014;9:14–21.
-
- Zhao Q., Ma L., Jiang S., Lu H., Liu S., He Y., Strick N., Neamati N., Debnath A.K. Identification of N-phenyl-N′-(2, 2, 6, 6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

