Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells
- PMID: 26295306
- PMCID: PMC4694895
- DOI: 10.18632/oncotarget.4613
Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells
Abstract
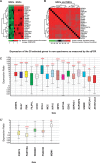
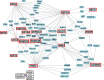
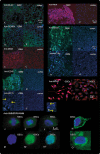
Glioblastoma (GBM) is both the most common and the most lethal primary brain tumor. It is thought that GBM stem cells (GSCs) are critically important in resistance to therapy. Therefore, there is a strong rationale to target these cells in order to develop new molecular therapies.To identify molecular targets in GSCs, we compared gene expression in GSCs to that in neural stem cells (NSCs) from the adult human brain, using microarrays. Bioinformatic filtering identified 20 genes (PBK/TOPK, CENPA, KIF15, DEPDC1, CDC6, DLG7/DLGAP5/HURP, KIF18A, EZH2, HMMR/RHAMM/CD168, NOL4, MPP6, MDM1, RAPGEF4, RHBDD1, FNDC3B, FILIP1L, MCC, ATXN7L4/ATXN7L1, P2RY5/LPAR6 and FAM118A) that were consistently expressed in GSC cultures and consistently not expressed in NSC cultures. The expression of these genes was confirmed in clinical samples (TCGA and REMBRANDT). The first nine genes were highly co-expressed in all GBM subtypes and were part of the same protein-protein interaction network. Furthermore, their combined up-regulation correlated negatively with patient survival in the mesenchymal GBM subtype. Using targeted proteomics and the COGNOSCENTE database we linked these genes to GBM signalling pathways.Nine genes: PBK, CENPA, KIF15, DEPDC1, CDC6, DLG7, KIF18A, EZH2 and HMMR should be further explored as targets for treatment of GBM.
Keywords: GBM; GSCs; glioblastoma; glioblastoma stem cells; therapeutic targeting.
Conflict of interest statement
None.
Figures





References
-
- Helseth R, Helseth E, Johannesen TB, Langberg CW, Lote K, Ronning P, Scheie D, Vik A, Meling TR. Overall survival, prognostic factors, and repeated surgery in a consecutive series of 516 patients with glioblastoma multiforme. Acta neurologica Scandinavica. 2010;122:159–167. - PubMed
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. Epub 2009 Mar 2009. - PubMed
-
- Moe MC, Varghese M, Danilov AI, Westerlund U, Ramm-Pettersen J, Brundin L, Svensson M, Berg-Johnsen J, Langmoen IA. Multipotent progenitor cells from the adult human brain: neurophysiological differentiation to mature neurons. Brain. 2005;128:2189–2199. - PubMed
-
- Moe MC, Westerlund U, Varghese M, Berg-Johnsen J, Svensson M, Langmoen IA. Development of neuronal networks from single stem cells harvested from the adult human brain. Neurosurgery. 2005;56:1182–1188. discussion 1188–1190. - PubMed
-
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nature medicine. 1998;4:1313–1317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

