The 26S proteasome is a multifaceted target for anti-cancer therapies
- PMID: 26295307
- PMCID: PMC4694792
- DOI: 10.18632/oncotarget.4619
The 26S proteasome is a multifaceted target for anti-cancer therapies
Abstract
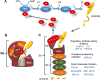
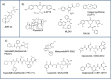
Proteasomes play a critical role in the fate of proteins that are involved in major cellular processes, including signal transduction, gene expression, cell cycle, replication, differentiation, immune response, cellular response to stress, etc. In contrast to non-specific degradation by lysosomes, proteasomes are highly selective and destroy only the proteins that are covalently labelled with small proteins, called ubiquitins. Importantly, many diseases, including neurodegenerative diseases and cancers, are intimately connected to the activity of proteasomes making them an important pharmacological target. Currently, the vast majority of inhibitors are aimed at blunting the proteolytic activities of proteasomes. However, recent achievements in solving structures of proteasomes at very high resolution provided opportunities to design new classes of small molecules that target other physiologically-important enzymatic activities of proteasomes, including the de-ubiquitinating one. This review attempts to catalog the information available to date about novel classes of proteasome inhibitors that may have important pharmacological ramifications.
Keywords: combined anti-cancer therapy; proteasome; proteasome inhibitors; ubiquitin-dependent proteolysis.
Conflict of interest statement
The authors state no conflict of interest.
Figures
References
-
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. - PMC - PubMed
-
- Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5046–5051. - PMC - PubMed
-
- Delcros JG, Floc'h MB, Prigent C, Arlot-Bonnemains Y. Proteasome inhibitors as therapeutic agents: current and future strategies. Current medicinal chemistry. 2003;10:479–503. - PubMed
-
- Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet. 2014 Dec;:pii. S0140-673660493-1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

