C/D box sRNA-guided 2'-O-methylation patterns of archaeal rRNA molecules
- PMID: 26296872
- PMCID: PMC4644070
- DOI: 10.1186/s12864-015-1839-z
C/D box sRNA-guided 2'-O-methylation patterns of archaeal rRNA molecules
Abstract
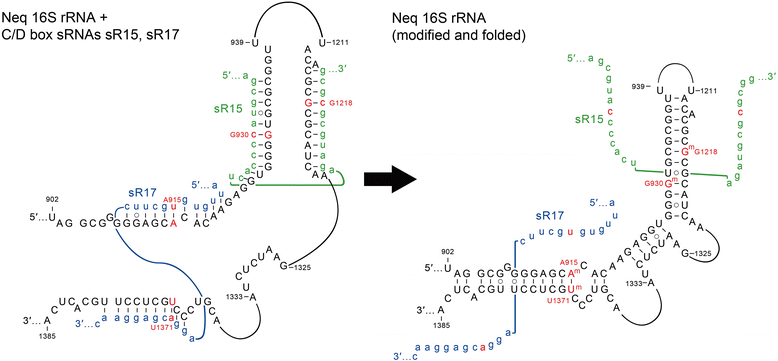
Background: In archaea and eukaryotes, ribonucleoprotein complexes containing small C/D box s(no)RNAs use base pair complementarity to target specific sites within ribosomal RNA for 2'-O-ribose methylation. These modifications aid in the folding and stabilization of nascent rRNA molecules and their assembly into ribosomal particles. The genomes of hyperthermophilic archaea encode large numbers of C/D box sRNA genes, suggesting an increased necessity for rRNA stabilization at extreme growth temperatures.
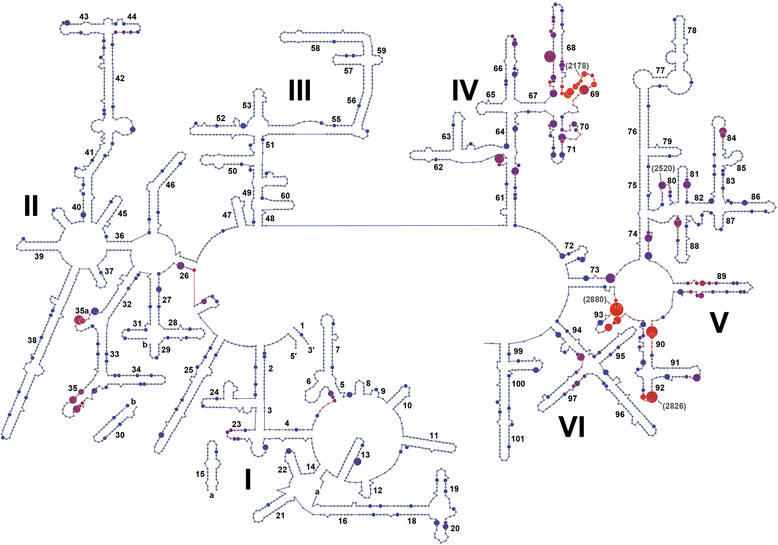
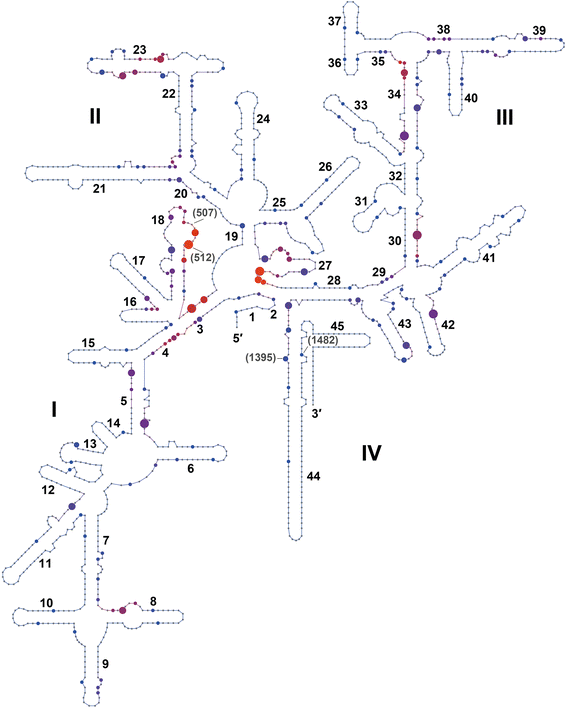
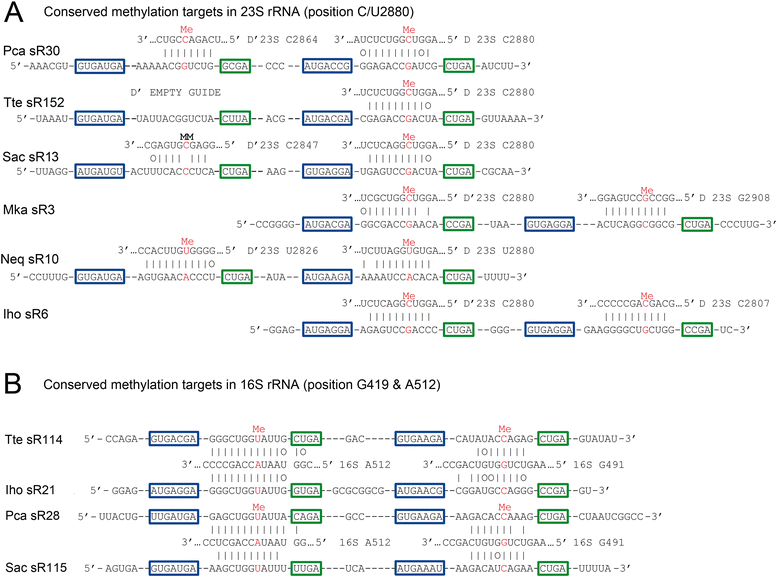
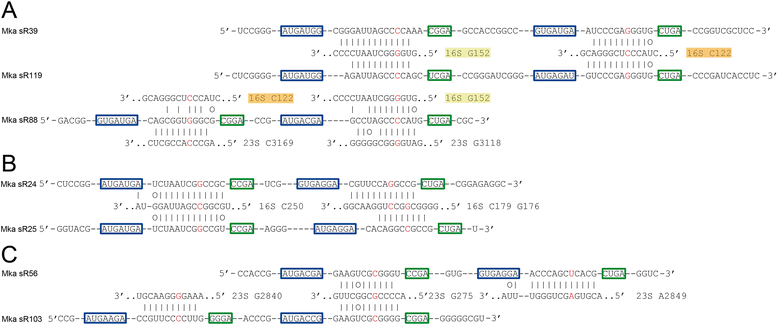
Results: We have identified the complete sets of C/D box sRNAs from seven archaea using RNA-Seq methodology. In total, 489 C/D box sRNAs were identified, each containing two guide regions. A combination of computational and manual analyses predicts 719 guide interactions with 16S and 23S rRNA molecules. This first pan-archaeal description of guide sequences identifies (i) modified rRNA nucleotides that are frequently conserved between species and (ii) regions within rRNA that are hotspots for 2'-O-methylation. Gene duplication, rearrangement, mutational drift and convergent evolution of sRNA genes and guide sequences were observed. In addition, several C/D box sRNAs were identified that use their two guides to target locations distant in the rRNA sequence but close in the secondary and tertiary structure. We propose that they act as RNA chaperones and facilitate complex folding events between distant sequences.
Conclusions: This pan-archaeal analysis of C/D box sRNA guide regions identified conserved patterns of rRNA 2'-O-methylation in archaea. The interaction between the sRNP complexes and the nascent rRNA facilitates proper folding and the methyl modifications stabilize higher order rRNA structure within the assembled ribosome.
Figures
Similar articles
-
Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling.J Mol Biol. 2006 Oct 6;362(5):1025-42. doi: 10.1016/j.jmb.2006.07.091. Epub 2006 Aug 4. J Mol Biol. 2006. PMID: 16949610
-
Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes.J Mol Biol. 2000 Apr 7;297(4):895-906. doi: 10.1006/jmbi.2000.3593. J Mol Biol. 2000. PMID: 10736225
-
Methylation guide RNA evolution in archaea: structure, function and genomic organization of 110 C/D box sRNA families across six Pyrobaculum species.Nucleic Acids Res. 2018 Jun 20;46(11):5678-5691. doi: 10.1093/nar/gky284. Nucleic Acids Res. 2018. PMID: 29771354 Free PMC article.
-
Small non-coding RNAs in Archaea.Curr Opin Microbiol. 2005 Dec;8(6):685-94. doi: 10.1016/j.mib.2005.10.013. Epub 2005 Oct 26. Curr Opin Microbiol. 2005. PMID: 16256421 Review.
-
The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA.Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):397-414. doi: 10.1002/wrna.117. Epub 2011 Nov 7. Wiley Interdiscip Rev RNA. 2012. PMID: 22065625 Review.
Cited by
-
Analysis of Expression Pattern of snoRNAs in Different Cancer Types with Machine Learning Algorithms.Int J Mol Sci. 2019 May 2;20(9):2185. doi: 10.3390/ijms20092185. Int J Mol Sci. 2019. PMID: 31052553 Free PMC article.
-
Box C/D guide RNAs recognize a maximum of 10 nt of substrates.Proc Natl Acad Sci U S A. 2016 Sep 27;113(39):10878-83. doi: 10.1073/pnas.1604872113. Epub 2016 Sep 13. Proc Natl Acad Sci U S A. 2016. PMID: 27625427 Free PMC article.
-
Archaeal fibrillarin-Nop5 heterodimer 2'-O-methylates RNA independently of the C/D guide RNP particle.RNA. 2017 Sep;23(9):1329-1337. doi: 10.1261/rna.059832.116. Epub 2017 Jun 2. RNA. 2017. PMID: 28576826 Free PMC article.
-
The extensive m5C epitranscriptome of Thermococcus kodakarensis is generated by a suite of RNA methyltransferases that support thermophily.Nat Commun. 2024 Aug 23;15(1):7272. doi: 10.1038/s41467-024-51410-w. Nat Commun. 2024. PMID: 39179532 Free PMC article.
-
RIP-Seq Suggests Translational Regulation by L7Ae in Archaea.mBio. 2017 Aug 1;8(4):e00730-17. doi: 10.1128/mBio.00730-17. mBio. 2017. PMID: 28765217 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

