Invasion of ovarian cancer cells is induced byPITX2-mediated activation of TGF-β and Activin-A
- PMID: 26298390
- PMCID: PMC4546816
- DOI: 10.1186/s12943-015-0433-y
Invasion of ovarian cancer cells is induced byPITX2-mediated activation of TGF-β and Activin-A
Abstract
Background: Most ovarian cancers are highly invasive in nature and the high burden of metastatic disease make them a leading cause of mortality among all gynaecological malignancies. The homeodomain transcription factor, PITX2 is associated with cancer in different tissues. Our previous studies demonstrated increased PITX2 expression in human ovarian tumours. Growing evidence linking activation of TGF-β pathway by homeodomain proteins prompted us to look for the possible involvement of this signalling pathway in PITX2-mediated progression of ovarian cancer.
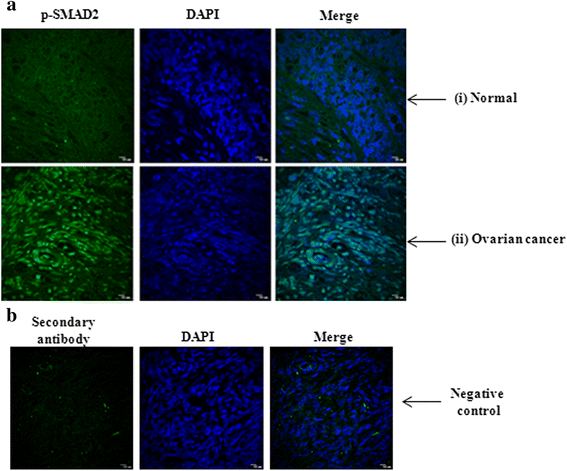
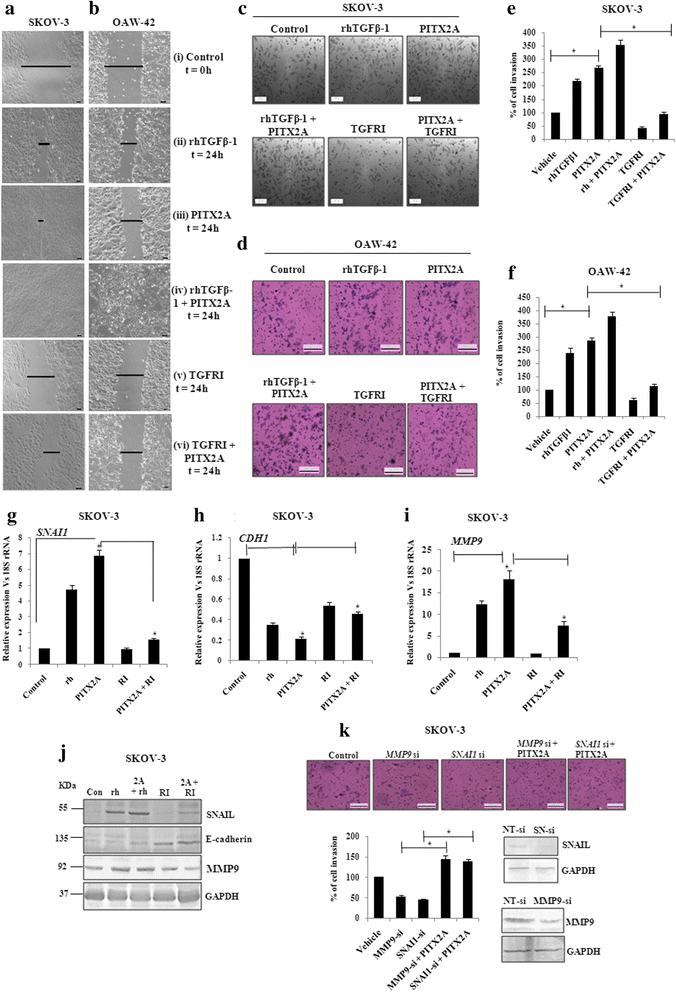
Methods: The status of TGF-β signalling in human ovarian tissues was assessed by immunohistochemistry. The expression level of TGFB/INHBA and other invasion-associated genes was measured by quantitative-PCR (Q-PCR) and Western Blot after transfection/treatments with clones/reagents in normal/cancer cells. The physiological effect of PITX2 on invasion/motility was checked by matrigel invasion and wound healing assay. The PITX2- and activin-induced epithelial-mesenchymal transition (EMT) was evaluated by Q-PCR of respective markers and confocal/phase-contrast imaging of cells.
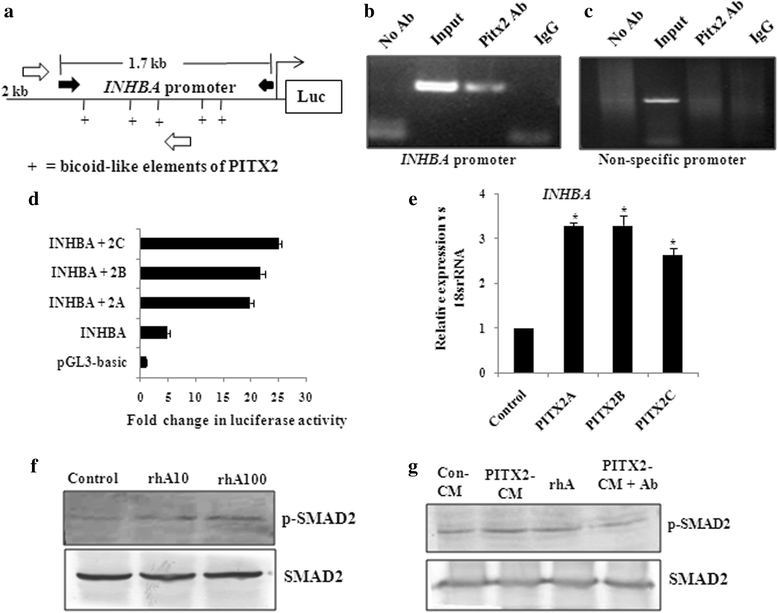
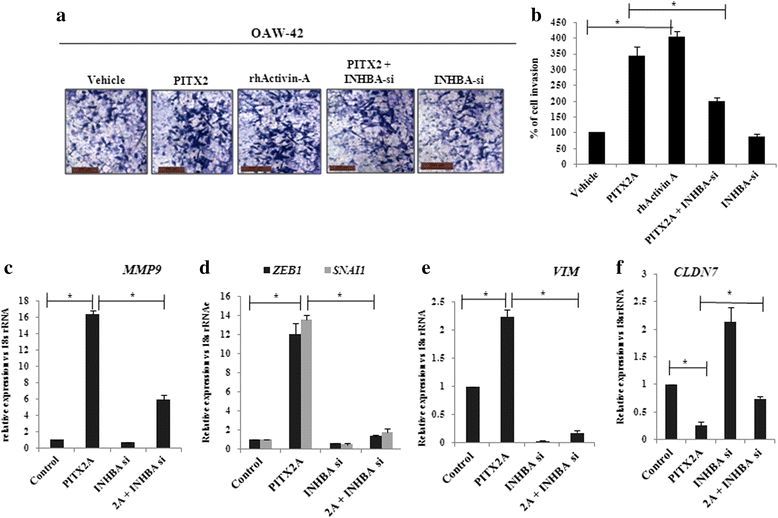
Results: Human ovarian tumours showed enhanced TGF-β signalling. Our study uncovers the PITX2-induced expression of TGFB1/2/3 as well as INHBA genes (p < 0.01) followed by SMAD2/3-dependent TGF-β signalling pathway. PITX2-induced TGF-β pathway regulated the expression of invasion-associated genes, SNAI1, CDH1 and MMP9 (p < 0.01) that accounted for enhanced motility/invasion of ovarian cancers. Snail and MMP9 acted as important mediators of PITX2-induced invasiveness of ovarian cancer cells. PITX2 over-expression resulted in loss of epithelial markers (p < 0.01) and gain of mesenchymal markers (p < 0.01) that contributed significantly to ovarian oncogenesis. PITX2-induced INHBA expression (p < 0.01) contributed to EMT in both normal and ovarian cancer cells.
Conclusions: Overall, our findings suggest a significant contributory role of PITX2 in promoting invasive behaviour of ovarian cancer cells through up-regulation of TGFB/INHBA. We have also identified the previously unknown involvement of activin-A in promoting EMT. Our work provides novel mechanistic insights into the invasive behavior of ovarian cancer cells. The extension of this study have the potential for therapeutic applications in future.
Figures



References
-
- Akhurst RJ, Derynck R. TGF-beta signaling in cancer-a double-edged sword. Trends Cell Biol. 2001;11:S44–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

