Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs
- PMID: 26299432
- PMCID: PMC4589543
- DOI: 10.1016/j.chom.2015.07.006
Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs
Abstract
Bacterial lineages that chronically infect cystic fibrosis (CF) patients genetically diversify during infection. However, the mechanisms driving diversification are unknown. By dissecting ten CF lung pairs and studying ∼12,000 regional isolates, we were able to investigate whether clonally related Pseudomonas aeruginosa inhabiting different lung regions evolve independently and differ functionally. Phylogenetic analysis of genome sequences showed that regional isolation of P. aeruginosa drives divergent evolution. We investigated the consequences of regional evolution by studying isolates from mildly and severely diseased lung regions and found evolved differences in bacterial nutritional requirements, host defense and antibiotic resistance, and virulence due to hyperactivity of the type 3 secretion system. These findings suggest that bacterial intermixing is limited in CF lungs and that regional selective pressures may markedly differ. The findings also may explain how specialized bacterial variants arise during infection and raise the possibility that pathogen diversification occurs in other chronic infections characterized by spatially heterogeneous conditions.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
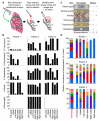
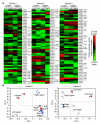


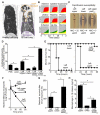

Comment in
-
Bacteria in the CF Lung: Isolation Drives Diversity.Cell Host Microbe. 2015 Sep 9;18(3):268-9. doi: 10.1016/j.chom.2015.08.013. Cell Host Microbe. 2015. PMID: 26355211
-
Bacterial evolution: To divide and conquer.Nat Rev Microbiol. 2015 Oct;13(10):602. doi: 10.1038/nrmicro3556. Nat Rev Microbiol. 2015. PMID: 26373369 No abstract available.
References
-
- Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 2001;183:444–452. - PubMed
-
- Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tummler B. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ. Microbiol. 2011;13:1690–1704. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

