Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses
- PMID: 26299452
- PMCID: PMC5026297
- DOI: 10.1016/j.cmet.2015.07.020
Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses
Abstract
The endolysosomal system is critical for the maintenance of cellular homeostasis. However, how endolysosomal compartment is regulated by mitochondrial function is largely unknown. We have generated a mouse model with defective mitochondrial function in CD4(+) T lymphocytes by genetic deletion of the mitochondrial transcription factor A (Tfam). Mitochondrial respiration deficiency impairs lysosome function, promotes p62 and sphingomyelin accumulation, and disrupts endolysosomal trafficking pathways and autophagy, thus linking a primary mitochondrial dysfunction to a lysosomal storage disorder. The impaired lysosome function in Tfam-deficient cells subverts T cell differentiation toward proinflammatory subsets and exacerbates the in vivo inflammatory response. Restoration of NAD(+) levels improves lysosome function and corrects the inflammatory defects in Tfam-deficient T cells. Our results uncover a mechanism by which mitochondria regulate lysosome function to preserve T cell differentiation and effector functions, and identify strategies for intervention in mitochondrial-related diseases.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

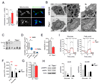
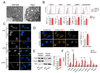

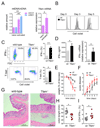
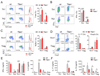
References
-
- Acin-Perez R, Carrascoso I, Baixauli F, Roche-Molina M, Latorre-Pellicer A, Fernandez-Silva P, Mittelbrunn M, Sanchez-Madrid F, Perez-Martos A, Lowell CA, et al. ROS-Triggered Phosphorylation of Complex II by Fgr Kinase Regulates Cellular Adaptation to Fuel Use. Cell Metab. 2014;19:1020–1033. - PMC - PubMed
-
- Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol. 2014;24:393–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

