Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries
- PMID: 26299473
- PMCID: PMC4560602
- DOI: 10.1016/j.neuron.2015.07.011
Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries
Abstract
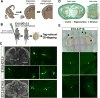
The medial ganglionic eminence (MGE) gives rise to the majority of mouse forebrain interneurons. Here, we examine the lineage relationship among MGE-derived interneurons using a replication-defective retroviral library containing a highly diverse set of DNA barcodes. Recovering the barcodes from the mature progeny of infected progenitor cells enabled us to unambiguously determine their respective lineal relationship. We found that clonal dispersion occurs across large areas of the brain and is not restricted by anatomical divisions. As such, sibling interneurons can populate the cortex, hippocampus striatum, and globus pallidus. The majority of interneurons appeared to be generated from asymmetric divisions of MGE progenitor cells, followed by symmetric divisions within the subventricular zone. Altogether, our findings uncover that lineage relationships do not appear to determine interneuron allocation to particular regions. As such, it is likely that clonally related interneurons have considerable flexibility as to the particular forebrain circuits to which they can contribute.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
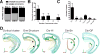


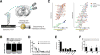
References
-
- Bates P, Young JAT, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

