Cardiolipin Interactions with Proteins
- PMID: 26300339
- PMCID: PMC4576322
- DOI: 10.1016/j.bpj.2015.07.034
Cardiolipin Interactions with Proteins
Abstract
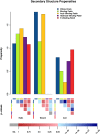
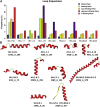
Cardiolipins (CL) represent unique phospholipids of bacteria and eukaryotic mitochondria with four acyl chains and two phosphate groups that have been implicated in numerous functions from energy metabolism to apoptosis. Many proteins are known to interact with CL, and several cocrystal structures of protein-CL complexes exist. In this work, we describe the collection of the first systematic and, to the best of our knowledge, the comprehensive gold standard data set of all known CL-binding proteins. There are 62 proteins in this data set, 21 of which have nonredundant crystal structures with bound CL molecules available. Using binding patch analysis of amino acid frequencies, secondary structures and loop supersecondary structures considering phosphate and acyl chain binding regions together and separately, we gained a detailed understanding of the general structural and dynamic features involved in CL binding to proteins. Exhaustive docking of CL to all known structures of proteins experimentally shown to interact with CL demonstrated the validity of the docking approach, and provides a rich source of information for experimentalists who may wish to validate predictions.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

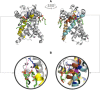

References
-
- Daum G. Lipids of mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. - PubMed
-
- Schlame M., Brody S., Hostetler K.Y. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur. J. Biochem. 1993;212:727–735. - PubMed
-
- Belikova N.A., Tyurina Y.Y., Kagan V.E. Heterolytic reduction of fatty acid hydroperoxides by cytochrome c/cardiolipin complexes: antioxidant function in mitochondria. J. Am. Chem. Soc. 2009;131:11288–11289. - PubMed
-
- Xu Y., Kelley R.I., Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 2003;278:51380–51385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

