Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes
- PMID: 26301496
- PMCID: PMC4780747
- DOI: 10.1038/ng.3385
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes
Abstract
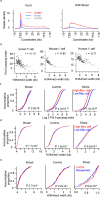
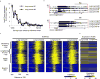
Tumor suppressors are mostly defined by inactivating mutations in tumors, yet little is known about their epigenetic features in normal cells. Through integrative analysis of 1,134 genome-wide epigenetic profiles, mutations from >8,200 tumor-normal pairs and our experimental data from clinical samples, we discovered broad peaks for trimethylation of histone H3 at lysine 4 (H3K4me3; wider than 4 kb) as the first epigenetic signature for tumor suppressors in normal cells. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity, which together lead to exceptionally high gene expression, and is distinct from other broad epigenetic features, such as super-enhancers. Genes with broad H3K4me3 peaks conserved across normal cells may represent pan-cancer tumor suppressors, such as TP53 and PTEN, whereas genes with cell type-specific broad H3K4me3 peaks may represent cell identity genes and cell type-specific tumor suppressors. Furthermore, widespread shortening of broad H3K4me3 peaks in cancers is associated with repression of tumor suppressors. Thus, the broad H3K4me3 epigenetic signature provides mutation-independent information for the discovery and characterization of new tumor suppressors.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interest that might influence the performance or presentation of the work described in this manuscript.
Figures






References
-
- Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012:1–5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

