Genetic association analyses highlight biological pathways underlying mitral valve prolapse
- PMID: 26301497
- PMCID: PMC4773907
- DOI: 10.1038/ng.3383
Genetic association analyses highlight biological pathways underlying mitral valve prolapse
Abstract
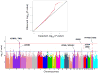
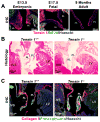
Nonsyndromic mitral valve prolapse (MVP) is a common degenerative cardiac valvulopathy of unknown etiology that predisposes to mitral regurgitation, heart failure and sudden death. Previous family and pathophysiological studies suggest a complex pattern of inheritance. We performed a meta-analysis of 2 genome-wide association studies in 1,412 MVP cases and 2,439 controls. We identified 6 loci, which we replicated in 1,422 cases and 6,779 controls, and provide functional evidence for candidate genes. We highlight LMCD1 (LIM and cysteine-rich domains 1), which encodes a transcription factor and for which morpholino knockdown of the ortholog in zebrafish resulted in atrioventricular valve regurgitation. A similar zebrafish phenotype was obtained with knockdown of the ortholog of TNS1, which encodes tensin 1, a focal adhesion protein involved in cytoskeleton organization. We also showed expression of tensin 1 during valve morphogenesis and describe enlarged posterior mitral leaflets in Tns1(-/-) mice. This study identifies the first risk loci for MVP and suggests new mechanisms involved in mitral valve regurgitation, the most common indication for mitral valve repair.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Freed LA, et al. Mitral valve prolapse in the general population: the benign nature of echocardiographic features in the Framingham Heart Study. J Am Coll Cardiol. 2002;40:1298–304. - PubMed
-
- Nesta F, et al. New locus for autosomal dominant mitral valve prolapse on chromosome 13: clinical insights from genetic studies. Circulation. 2005;112:2022–30. - PubMed
Publication types
MeSH terms
Grants and funding
- K24 HL067434/HL/NHLBI NIH HHS/United States
- R01 HL127692/HL/NHLBI NIH HHS/United States
- R01 HL104156/HL/NHLBI NIH HHS/United States
- U01 HL065962/HL/NHLBI NIH HHS/United States
- MOP-102737/CAPMC/ CIHR/Canada
- HL109506/HL/NHLBI NIH HHS/United States
- MOP-114997/CAPMC/ CIHR/Canada
- R01 HL109004/HL/NHLBI NIH HHS/United States
- MOP-137058/CAPMC/ CIHR/Canada
- MOP-126072/CAPMC/ CIHR/Canada
- C06 RR018823/RR/NCRR NIH HHS/United States
- 1P30 GM103342/GM/NIGMS NIH HHS/United States
- HL092577/HL/NHLBI NIH HHS/United States
- HL104156/HL/NHLBI NIH HHS/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- R01 HL109506/HL/NHLBI NIH HHS/United States
- R01-HL127692/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- K24HL105780/HL/NHLBI NIH HHS/United States
- 8P20 GM103444-07/GM/NIGMS NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- HL065962/HL/NHLBI NIH HHS/United States
- K24 HL67434/HL/NHLBI NIH HHS/United States
- R01 HL128099/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- R01 HL72265/HL/NHLBI NIH HHS/United States
- R01 HL072265/HL/NHLBI NIH HHS/United States
- MOP-102481/CAPMC/ CIHR/Canada
- K23 HL116652/HL/NHLBI NIH HHS/United States
- R01-HL33756/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

