Cancer Cell Growth Is Differentially Affected by Constitutive Activation of NRF2 by KEAP1 Deletion and Pharmacological Activation of NRF2 by the Synthetic Triterpenoid, RTA 405
- PMID: 26301506
- PMCID: PMC4547720
- DOI: 10.1371/journal.pone.0135257
Cancer Cell Growth Is Differentially Affected by Constitutive Activation of NRF2 by KEAP1 Deletion and Pharmacological Activation of NRF2 by the Synthetic Triterpenoid, RTA 405
Abstract
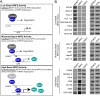
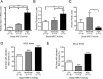
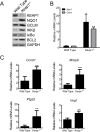
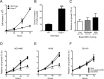
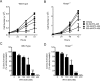
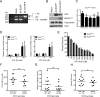
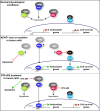
Synthetic triterpenoids are antioxidant inflammation modulators (AIMs) that exhibit broad anticancer activity. AIMs bind to KEAP1 and inhibit its ability to promote NRF2 degradation. As a result, NRF2 increases transcription of genes that restore redox balance and reduce inflammation. AIMs inhibit tumor growth and metastasis by increasing NRF2 activity in the tumor microenvironment and by modulating the activity of oncogenic signaling pathways, including NF-κB, in tumor cells. Accumulating evidence suggests that KEAP1 loss or mutation--which results in high levels of sustained NRF2 activity--may promote cancer growth and increase chemoresistance. Loss of KEAP1 also increases the levels of other oncogenic proteins, including IKKβ and BCL2. The apparent survival advantage provided to some tumor cells by loss of functional KEAP1 raises the question of whether pharmacological inhibition of KEAP1 could promote tumor growth. To address this issue, we characterized the basal levels of KEAP1 and NRF2 in a panel of human tumor cell lines and profiled the activity of an AIM, RTA 405. We found that in tumor cell lines with low or mutant KEAP1, and in Keap1-/- murine embryonic fibroblasts, multiple KEAP1 targets including NRF2, IKKβ, and BCL2 were elevated. Keap1-/- murine embryonic fibroblasts also had higher rates of proliferation and colony formation than their wild-type counterparts. In cells with functional KEAP1, RTA 405 increased NRF2 levels, but not IKKβ or BCL2 levels, and did not increase cell proliferation or survival. Moreover, RTA 405 inhibited growth at similar concentrations in cells with different basal NRF2 activity levels and in cells with wild-type or mutant KRAS. Finally, pre-treatment with RTA 405 did not protect tumor cells from doxorubicin- or cisplatin-mediated growth inhibition. Collectively, these data demonstrate that pharmacological activation of NRF2 by AIMs is distinct from genetic activation and does not provide a growth or survival advantage to tumor cells.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

