Predicting effects of noncoding variants with deep learning-based sequence model
- PMID: 26301843
- PMCID: PMC4768299
- DOI: 10.1038/nmeth.3547
Predicting effects of noncoding variants with deep learning-based sequence model
Abstract
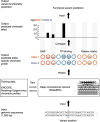
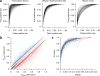
Identifying functional effects of noncoding variants is a major challenge in human genetics. To predict the noncoding-variant effects de novo from sequence, we developed a deep learning-based algorithmic framework, DeepSEA (http://deepsea.princeton.edu/), that directly learns a regulatory sequence code from large-scale chromatin-profiling data, enabling prediction of chromatin effects of sequence alterations with single-nucleotide sensitivity. We further used this capability to improve prioritization of functional variants including expression quantitative trait loci (eQTLs) and disease-associated variants.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Diving deeper to predict noncoding sequence function.Nat Methods. 2015 Oct;12(10):925-6. doi: 10.1038/nmeth.3604. Nat Methods. 2015. PMID: 26418766 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

