Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome
- PMID: 26301997
- PMCID: PMC4560640
- DOI: 10.1038/nsmb.3075
Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome
Abstract
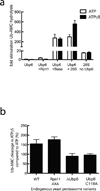
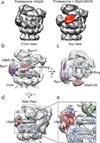
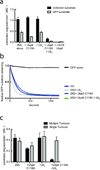
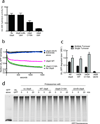
Substrates are targeted for proteasomal degradation through the attachment of ubiquitin chains that need to be removed by proteasomal deubiquitinases before substrate processing. In budding yeast, the deubiquitinase Ubp6 trims ubiquitin chains and affects substrate processing by the proteasome, but the underlying mechanisms and the location of Ubp6 within the holoenzyme have been elusive. Here we show that Ubp6 activity strongly responds to interactions with the base ATPase and the conformational state of the proteasome. Electron microscopy analyses reveal that ubiquitin-bound Ubp6 contacts the N ring and AAA+ ring of the ATPase hexamer and is in proximity to the deubiquitinase Rpn11. Ubiquitin-bound Ubp6 inhibits substrate deubiquitination by Rpn11, stabilizes the substrate-engaged conformation of the proteasome and allosterically interferes with the engagement of a subsequent substrate. Ubp6 may thus act as a ubiquitin-dependent 'timer' to coordinate individual processing steps at the proteasome and modulate substrate degradation.
Figures


Comment in
-
A timer to coordinate substrate processing by the 26S proteasome.Nat Struct Mol Biol. 2015 Sep;22(9):652-3. doi: 10.1038/nsmb.3085. Nat Struct Mol Biol. 2015. PMID: 26333712 No abstract available.
References
-
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. - PubMed
-
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 2007;35:12–17. - PubMed
-
- Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. - PubMed
-
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

