Trichodysplasia spinulosa-Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- PMID: 26302170
- PMCID: PMC4547793
- DOI: 10.1371/journal.ppat.1005112
Trichodysplasia spinulosa-Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
Abstract
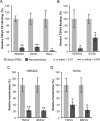
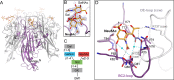
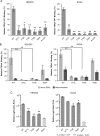
Trichodysplasia spinulosa-associated Polyomavirus (TSPyV) was isolated from a patient suffering from trichodysplasia spinulosa, a skin disease that can appear in severely immunocompromised patients. While TSPyV is one of the five members of the polyomavirus family that are directly linked to a human disease, details about molecular recognition events, the viral entry pathway, and intracellular trafficking events during TSPyV infection remain unknown. Here we have used a structure-function approach to shed light on the first steps of TSPyV infection. We established by cell binding and pseudovirus infection studies that TSPyV interacts with sialic acids during attachment and/or entry. Subsequently, we solved high-resolution X-ray structures of the major capsid protein VP1 of TSPyV in complex with three different glycans, the branched GM1 glycan, and the linear trisaccharides α2,3- and α2,6-sialyllactose. The terminal sialic acid of all three glycans is engaged in a unique binding site on TSPyV VP1, which is positioned about 18 Å from established sialic acid binding sites of other polyomaviruses. Structure-based mutagenesis of sialic acid-binding residues leads to reduction in cell attachment and pseudovirus infection, demonstrating the physiological relevance of the TSPyV VP1-glycan interaction. Furthermore, treatments of cells with inhibitors of N-, O-linked glycosylation, and glycosphingolipid synthesis suggest that glycolipids play an important role during TSPyV infection. Our findings elucidate the first molecular recognition events of cellular infection with TSPyV and demonstrate that receptor recognition by polyomaviruses is highly variable not only in interactions with sialic acid itself, but also in the location of the binding site.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
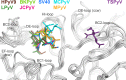
Similar articles
-
Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.mBio. 2020 Jul 28;11(4):e00745-20. doi: 10.1128/mBio.00745-20. mBio. 2020. PMID: 32723915 Free PMC article.
-
Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic acid binding site.J Virol. 2014 Sep;88(18):10831-9. doi: 10.1128/JVI.01084-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008942 Free PMC article.
-
Production and characterization of monoclonal antibodies specific for major capsid VP1 protein of trichodysplasia spinulosa-associated polyomavirus.Microbiol Immunol. 2018 Dec;62(12):763-773. doi: 10.1111/1348-0421.12662. Microbiol Immunol. 2018. PMID: 30537287
-
The trichodysplasia spinulosa-associated polyomavirus: virological background and clinical implications.APMIS. 2013 Aug;121(8):770-82. doi: 10.1111/apm.12092. Epub 2013 Apr 18. APMIS. 2013. PMID: 23593936 Review.
-
Pediatric trichodysplasia spinulosa: A report of 2 cases and review of the literature.Pediatr Dermatol. 2020 Nov;37(6):1023-1029. doi: 10.1111/pde.14323. Epub 2020 Aug 12. Pediatr Dermatol. 2020. PMID: 32785992 Review.
Cited by
-
Carbohydrates: Binding Sites and Potential Drug Targets for Neural-Affecting Pathogens.Adv Neurobiol. 2023;29:449-477. doi: 10.1007/978-3-031-12390-0_15. Adv Neurobiol. 2023. PMID: 36255684
-
Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.mBio. 2020 Jul 28;11(4):e00745-20. doi: 10.1128/mBio.00745-20. mBio. 2020. PMID: 32723915 Free PMC article.
-
Structural Insight into Non-Enveloped Virus Binding to Glycosaminoglycan Receptors: A Review.Viruses. 2021 Apr 29;13(5):800. doi: 10.3390/v13050800. Viruses. 2021. PMID: 33946963 Free PMC article. Review.
-
Agglutination of Human Polyomaviruses by Using a Tetravalent Glycocluster as a Cross-Linker.ACS Omega. 2020 Aug 18;5(34):21940-21947. doi: 10.1021/acsomega.0c03269. eCollection 2020 Sep 1. ACS Omega. 2020. PMID: 32905316 Free PMC article.
-
The Symmetry of Viral Sialic Acid Binding Sites-Implications for Antiviral Strategies.Viruses. 2019 Oct 14;11(10):947. doi: 10.3390/v11100947. Viruses. 2019. PMID: 31615155 Free PMC article. Review.
References
-
- Haycox CL, Kim S, Fleckman P, Smith LT, Piepkorn M, et al. (1999) Trichodysplasia spinulosa—a newly described folliculocentric viral infection in an immunocompromised host. The journal of investigative dermatology Symposium proceedings / the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research 4: 268–271. - PubMed
-
- Heaphy MR Jr., Shamma HN, Hickmann M, White MJ (2004) Cyclosporine-induced folliculodystrophy. Journal of the American Academy of Dermatology 50: 310–315. - PubMed
-
- Sperling LC, Tomaszewski MM, Thomas DA (2004) Viral-associated trichodysplasia in patients who are immunocompromised. Journal of the American Academy of Dermatology 50: 318–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

