Phenotypic states become increasingly sensitive to perturbations near a bifurcation in a synthetic gene network
- PMID: 26302311
- PMCID: PMC4547091
- DOI: 10.7554/eLife.07935
Phenotypic states become increasingly sensitive to perturbations near a bifurcation in a synthetic gene network
Abstract
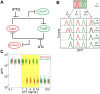
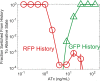
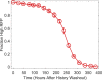
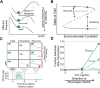
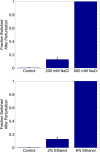
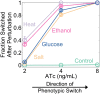
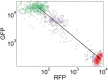
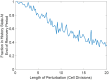
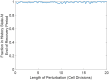
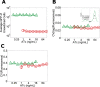
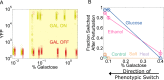
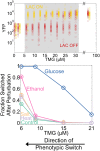
Microorganisms often exhibit a history-dependent phenotypic response after exposure to a stimulus which can be imperative for proper function. However, cells frequently experience unexpected environmental perturbations that might induce phenotypic switching. How cells maintain phenotypic states in the face of environmental fluctuations remains an open question. Here, we use environmental perturbations to characterize the resilience of phenotypic states in a synthetic gene network near a critical transition. We find that far from the critical transition an environmental perturbation may induce little to no phenotypic switching, whereas close to the critical transition the same perturbation can cause many cells to switch phenotypic states. This loss of resilience was observed for perturbations that interact directly with the gene circuit as well as for a variety of generic perturbations-such as salt, ethanol, or temperature shocks-that alter the state of the cell more broadly. We obtain qualitatively similar findings in natural gene circuits, such as the yeast GAL network. Our findings illustrate how phenotypic memory can become destabilized by environmental variability near a critical transition.
Keywords: E. coli; S. cerevisiae; cell memory; computational biology; critical transitions; flow cytometry; resilience; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

