Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex
- PMID: 26303005
- PMCID: PMC4780368
- DOI: 10.1016/j.virol.2015.07.018
Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex
Abstract
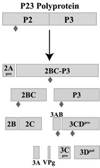
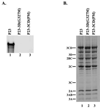
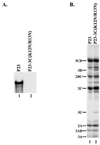
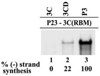
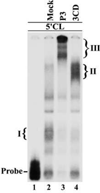
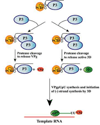
The differential use of protein precursors and their products is a key strategy used during poliovirus replication. To characterize the role of protein precursors during replication, we examined the complementation profiles of mutants that inhibited 3D polymerase or 3C-RNA binding activity. We showed that 3D entered the replication complex in the form of its precursor, P3 (or 3CD), and was cleaved to release active 3D polymerase. Furthermore, our results showed that P3 is the preferred precursor that binds to the 5'CL. Using reciprocal complementation assays, we showed that one molecule of P3 binds the 5'CL and that a second molecule of P3 provides 3D. In addition, we showed that a second molecule of P3 served as the VPg provider. These results support a model in which P3 binds to the 5'CL and recruits additional molecules of P3, which are cleaved to release either 3D or VPg to initiate RNA replication.
Keywords: (–) Strand RNA; 3C-RNA binding activity; 3D polymerase; 5′Cloverleaf (5′CL); Poliovirus (PV); Poliovirus 3CD; Poliovirus P3; RNA replication complex; Reciprocal complementation; VPg.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
 ).
).





Similar articles
-
Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB.J Virol. 2007 Jun;81(11):5669-84. doi: 10.1128/JVI.02350-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360746 Free PMC article.
-
The 5'CL-PCBP RNP complex, 3' poly(A) tail and 2A(pro) are required for optimal translation of poliovirus RNA.Virology. 2010 Feb 5;397(1):14-22. doi: 10.1016/j.virol.2009.11.006. Epub 2009 Nov 27. Virology. 2010. PMID: 19945132 Free PMC article.
-
Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation.J Virol. 2000 Nov;74(22):10371-80. doi: 10.1128/jvi.74.22.10371-10380.2000. J Virol. 2000. PMID: 11044081 Free PMC article.
-
Non-template functions of viral RNA in picornavirus replication.Curr Opin Virol. 2011 Nov;1(5):339-46. doi: 10.1016/j.coviro.2011.09.005. Curr Opin Virol. 2011. PMID: 22140418 Free PMC article. Review.
-
Expanding knowledge of P3 proteins in the poliovirus lifecycle.Future Microbiol. 2010 Jun;5(6):867-81. doi: 10.2217/fmb.10.40. Future Microbiol. 2010. PMID: 20521933 Free PMC article. Review.
Cited by
-
Processing of the 3C/D Region of the Deformed Wing Virus (DWV).Viruses. 2023 Nov 29;15(12):2344. doi: 10.3390/v15122344. Viruses. 2023. PMID: 38140585 Free PMC article.
-
Insights into Polyprotein Processing and RNA-Protein Interactions in Foot-and-Mouth Disease Virus Genome Replication.J Virol. 2023 May 31;97(5):e0017123. doi: 10.1128/jvi.00171-23. Epub 2023 May 8. J Virol. 2023. PMID: 37154761 Free PMC article.
-
Enterovirus C recombination groups: RNA sequence similarity and the viral polymerase underpin sexual replication mechanisms.J Virol. 2025 Jul 22;99(7):e0043425. doi: 10.1128/jvi.00434-25. Epub 2025 Jun 24. J Virol. 2025. PMID: 40552818 Free PMC article.
-
The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3Cpro and 3Dpol in Functionally Relevant Regions.Viruses. 2021 Mar 9;13(3):442. doi: 10.3390/v13030442. Viruses. 2021. PMID: 33803479 Free PMC article.
-
Both cis and trans Activities of Foot-and-Mouth Disease Virus 3D Polymerase Are Essential for Viral RNA Replication.J Virol. 2016 Jul 11;90(15):6864-6883. doi: 10.1128/JVI.00469-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194768 Free PMC article.
References
-
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990a;63:369–380. - PubMed
-
- Barton DJ, Morasco BJ, Flanegan JB. Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

