The expanded genetic alphabet
- PMID: 26304162
- PMCID: PMC4798003
- DOI: 10.1002/anie.201502890
The expanded genetic alphabet
Abstract
All biological information, since the last common ancestor of all life on Earth, has been encoded by a genetic alphabet consisting of only four nucleotides that form two base pairs. Long-standing efforts to develop two synthetic nucleotides that form a third, unnatural base pair (UBP) have recently yielded three promising candidates, one based on alternative hydrogen bonding, and two based on hydrophobic and packing forces. All three of these UBPs are replicated and transcribed with remarkable efficiency and fidelity, and the latter two thus demonstrate that hydrogen bonding is not unique in its ability to underlie the storage and retrieval of genetic information. This Review highlights these recent developments as well as the applications enabled by the UBPs, including the expansion of the evolution process to include new functionality and the creation of semi-synthetic life that stores increased information.
Keywords: DNA; base pairs; hydrophobicity; semi-synthetic organism; unnatural nucleotides.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

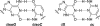

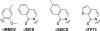

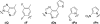

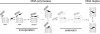

References
-
- Rich A. Horizons in Biochemistry. Academic Press; New York: 1962.
-
- Fa M, Radeghieri A, Henry AA, Romesberg FE. J. Am. Chem. Soc. 2004;126:1748–1754. - PubMed
-
- Veedu RN, Wengel J. Chem. Biodiver. 2010;7:536–542. - PubMed
-
- Chaput JC, Yu HY, Zhang S. Chem. Biol. 2012;19:1360–1371. - PubMed
-
- Pinheiro VB, Holliger P. Curr. Opin. Chem. Biol. 2012;16:245–252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

