LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations
- PMID: 26304545
- PMCID: PMC4787796
- DOI: 10.1093/nar/gkv803
LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations
Abstract
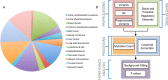
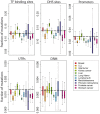
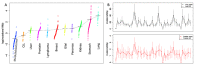
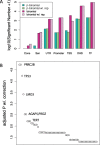
In cancer research, background models for mutation rates have been extensively calibrated in coding regions, leading to the identification of many driver genes, recurrently mutated more than expected. Noncoding regions are also associated with disease; however, background models for them have not been investigated in as much detail. This is partially due to limited noncoding functional annotation. Also, great mutation heterogeneity and potential correlations between neighboring sites give rise to substantial overdispersion in mutation count, resulting in problematic background rate estimation. Here, we address these issues with a new computational framework called LARVA. It integrates variants with a comprehensive set of noncoding functional elements, modeling the mutation counts of the elements with a β-binomial distribution to handle overdispersion. LARVA, moreover, uses regional genomic features such as replication timing to better estimate local mutation rates and mutational hotspots. We demonstrate LARVA's effectiveness on 760 whole-genome tumor sequences, showing that it identifies well-known noncoding drivers, such as mutations in the TERT promoter. Furthermore, LARVA highlights several novel highly mutated regulatory sites that could potentially be noncoding drivers. We make LARVA available as a software tool and release our highly mutated annotations as an online resource (larva.gersteinlab.org).
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Almasy L., Dyer T.D., Peralta J.M., Jun G., Wood A.R., Fuchsberger C., Almeida M.A., Kent J.W., Fowler S., Blackwell T.W., et al. Data for Genetic Analysis Workshop 18: human whole genome sequence, blood pressure, and simulated phenotypes in extended pedigrees. BMC Proc. 2014;8:S2–S10. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

