Transcription errors induce proteotoxic stress and shorten cellular lifespan
- PMID: 26304740
- PMCID: PMC4684168
- DOI: 10.1038/ncomms9065
Transcription errors induce proteotoxic stress and shorten cellular lifespan
Erratum in
-
Corrigendum: Transcription errors induce proteotoxic stress and shorten cellular lifespan.Nat Commun. 2015 Oct 14;6:8738. doi: 10.1038/ncomms9738. Nat Commun. 2015. PMID: 26465398 No abstract available.
Abstract
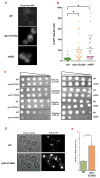
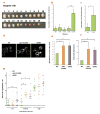
Transcription errors occur in all living cells; however, it is unknown how these errors affect cellular health. To answer this question, we monitor yeast cells that are genetically engineered to display error-prone transcription. We discover that these cells suffer from a profound loss in proteostasis, which sensitizes them to the expression of genes that are associated with protein-folding diseases in humans; thus, transcription errors represent a new molecular mechanism by which cells can acquire disease phenotypes. We further find that the error rate of transcription increases as cells age, suggesting that transcription errors affect proteostasis particularly in aging cells. Accordingly, transcription errors accelerate the aggregation of a peptide that is implicated in Alzheimer's disease, and shorten the lifespan of cells. These experiments reveal a previously unappreciated role for transcriptional fidelity in cellular health and aging.
Figures


References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. - PubMed
-
- Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. Faseb J. 2001;15(2):322–332. - PubMed
-
- Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. - PubMed
-
- Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284(5411):159–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008570/GM/NIGMS NIH HHS/United States
- GM092741/GM/NIGMS NIH HHS/United States
- P01 AG001751/AG/NIA NIH HHS/United States
- R01 GM056981/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01AG039390/AG/NIA NIH HHS/United States
- T32AG000057/AG/NIA NIH HHS/United States
- NIA 11006596/PHS HHS/United States
- R01 GM092741/GM/NIGMS NIH HHS/United States
- R01 GM079480/GM/NIGMS NIH HHS/United States
- GM 080294/GM/NIGMS NIH HHS/United States
- T32ES007032/ES/NIEHS NIH HHS/United States
- R01GM56981/GM/NIGMS NIH HHS/United States
- R01 GM080294/GM/NIGMS NIH HHS/United States
- R01 GM067785/GM/NIGMS NIH HHS/United States
- P30 AG013280/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

