New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae
- PMID: 26305040
- PMCID: PMC4715497
- DOI: 10.1002/yea.3098
New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology is an important tool for genome editing because the Cas9 endonuclease can induce targeted DNA double-strand breaks. Targeting of the DNA break is typically controlled by a single-guide RNA (sgRNA), a chimeric RNA containing a structural segment important for Cas9 binding and a 20mer guide sequence that hybridizes to the genomic DNA target. Previous studies have demonstrated that CRISPR-Cas9 technology can be used for efficient, marker-free genome editing in Saccharomyces cerevisiae. However, introducing the 20mer guide sequence into yeast sgRNA expression vectors often requires cloning procedures that are complex, time-consuming and/or expensive. To simplify this process, we have developed a new sgRNA expression cassette with internal restriction enzyme sites that permit rapid, directional cloning of 20mer guide sequences. Here we describe a flexible set of vectors based on this design for cloning and expressing sgRNAs (and Cas9) in yeast using different selectable markers. We anticipate that the Cas9-sgRNA expression vector with the URA3 selectable marker (pML104) will be particularly useful for genome editing in yeast, since the Cas9 machinery can be easily removed by counter-selection using 5-fluoro-orotic acid (5-FOA) following successful genome editing. The availability of new vectors that simplify and streamline the technical steps required for guide sequence cloning should help accelerate the use of CRISPR-Cas9 technology in yeast genome editing.
Keywords: CRISPR; Cas9; Saccharomyces cerevisiae; genome editing; guide RNA; plasmids.
Copyright © 2015 John Wiley & Sons, Ltd.
Figures

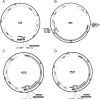
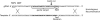
References
-
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H. Homology-Integrated CRISPR-Cas (HI-CRISPR) System for One-Step Multigene Disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:585–94. - PubMed
-
- Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–39. - PubMed
-
- Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
