Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia
- PMID: 26305651
- PMCID: PMC4621257
- DOI: 10.1001/jama.2015.9643
Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia
Abstract
Importance: Tests that predict outcomes for patients with acute myeloid leukemia (AML) are imprecise, especially for those with intermediate risk AML.
Objectives: To determine whether genomic approaches can provide novel prognostic information for adult patients with de novo AML.
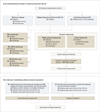
Design, setting, and participants: Whole-genome or exome sequencing was performed on samples obtained at disease presentation from 71 patients with AML (mean age, 50.8 years) treated with standard induction chemotherapy at a single site starting in March 2002, with follow-up through January 2015. In addition, deep digital sequencing was performed on paired diagnosis and remission samples from 50 patients (including 32 with intermediate-risk AML), approximately 30 days after successful induction therapy. Twenty-five of the 50 were from the cohort of 71 patients, and 25 were new, additional cases.
Exposures: Whole-genome or exome sequencing and targeted deep sequencing. Risk of identification based on genetic data.
Main outcomes and measures: Mutation patterns (including clearance of leukemia-associated variants after chemotherapy) and their association with event-free survival and overall survival.
Results: Analysis of comprehensive genomic data from the 71 patients did not improve outcome assessment over current standard-of-care metrics. In an analysis of 50 patients with both presentation and documented remission samples, 24 (48%) had persistent leukemia-associated mutations in at least 5% of bone marrow cells at remission. The 24 with persistent mutations had significantly reduced event-free and overall survival vs the 26 who cleared all mutations. Patients with intermediate cytogenetic risk profiles had similar findings. [table: see text].
Conclusions and relevance: The detection of persistent leukemia-associated mutations in at least 5% of bone marrow cells in day 30 remission samples was associated with a significantly increased risk of relapse, and reduced overall survival. These data suggest that this genomic approach may improve risk stratification for patients with AML.
Conflict of interest statement
Figures




Comment in
-
Next-Generation Sequencing and Detection of Minimal Residual Disease in Acute Myeloid Leukemia: Ready for Clinical Practice?JAMA. 2015 Aug 25;314(8):778-80. doi: 10.1001/jama.2015.9452. JAMA. 2015. PMID: 26305647 No abstract available.
-
Haematological cancer: Improving prediction of relapse risk.Nat Rev Clin Oncol. 2015 Oct;12(10):564. doi: 10.1038/nrclinonc.2015.156. Epub 2015 Sep 8. Nat Rev Clin Oncol. 2015. PMID: 26346846 No abstract available.
References
-
- Breems DA, Van Putten WL, Huijgens PC, et al. Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol. 2005;23(9):1969–1978. - PubMed
-
- Walter RB, Kantarjian HM, Huang X, et al. Effect of complete remission and responses less than complete remission on survival in acute myeloid leukemia: a combined Eastern Cooperative Oncology Group, Southwest Oncology Group, and M.D. Anderson Cancer Center Study. J Clin Oncol. 2010;28(10):1766–1771. - PMC - PubMed
-
- Byrd JC, Mrózek K, Dodge RK, et al. Cancer and Leukemia Group B (CALGB 8461). Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100(13):4325–4336. - PubMed
-
- Kihara R, Nagata Y, Kiyoi H, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28(8):1586–1595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

