Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira
- PMID: 26305944
- PMCID: PMC4568715
- DOI: 10.1073/pnas.1506533112
Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira
Abstract
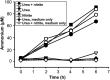
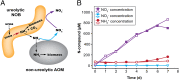
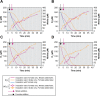
Nitrospira are a diverse group of nitrite-oxidizing bacteria and among the environmentally most widespread nitrifiers. However, they remain scarcely studied and mostly uncultured. Based on genomic and experimental data from Nitrospira moscoviensis representing the ubiquitous Nitrospira lineage II, we identified ecophysiological traits that contribute to the ecological success of Nitrospira. Unexpectedly, N. moscoviensis possesses genes coding for a urease and cleaves urea to ammonia and CO2. Ureolysis was not observed yet in nitrite oxidizers and enables N. moscoviensis to supply ammonia oxidizers lacking urease with ammonia from urea, which is fully nitrified by this consortium through reciprocal feeding. The presence of highly similar urease genes in Nitrospira lenta from activated sludge, in metagenomes from soils and freshwater habitats, and of other ureases in marine nitrite oxidizers, suggests a wide distribution of this extended interaction between ammonia and nitrite oxidizers, which enables nitrite-oxidizing bacteria to indirectly use urea as a source of energy. A soluble formate dehydrogenase lends additional ecophysiological flexibility and allows N. moscoviensis to use formate, with or without concomitant nitrite oxidation, using oxygen, nitrate, or both compounds as terminal electron acceptors. Compared with Nitrospira defluvii from lineage I, N. moscoviensis shares the Nitrospira core metabolism but shows substantial genomic dissimilarity including genes for adaptations to elevated oxygen concentrations. Reciprocal feeding and metabolic versatility, including the participation in different nitrogen cycling processes, likely are key factors for the niche partitioning, the ubiquity, and the high diversity of Nitrospira in natural and engineered ecosystems.
Keywords: Nitrospira; formate; genome; nitrification; urease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Kraft B, et al. Nitrogen cycling. The environmental controls that govern the end product of bacterial nitrate respiration. Science. 2014;345(6197):676–679. - PubMed
-
- Prosser JI. Soil nitrifiers and nitrification. In: Ward BB, Arp DJ, Klotz MG, editors. Nitrification. Am Soc Microbiol; Washington: 2011. pp. 347–383.
-
- Watson SW, Waterbury JB. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Mikrobiol. 1971;77:203–230.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

