Methods for analysis of size-exclusion chromatography-small-angle X-ray scattering and reconstruction of protein scattering
- PMID: 26306089
- PMCID: PMC4520288
- DOI: 10.1107/S1600576715010420
Methods for analysis of size-exclusion chromatography-small-angle X-ray scattering and reconstruction of protein scattering
Abstract
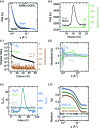
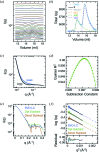
Size-exclusion chromatography in line with small-angle X-ray scattering (SEC-SAXS) has emerged as an important method for investigation of heterogeneous and self-associating systems, but presents specific challenges for data processing including buffer subtraction and analysis of overlapping peaks. This paper presents novel methods based on singular value decomposition (SVD) and Guinier-optimized linear combination (LC) to facilitate analysis of SEC-SAXS data sets and high-quality reconstruction of protein scattering directly from peak regions. It is shown that Guinier-optimized buffer subtraction can reduce common subtraction artifacts and that Guinier-optimized linear combination of significant SVD basis components improves signal-to-noise and allows reconstruction of protein scattering, even in the absence of matching buffer regions. In test cases with conventional SAXS data sets for cytochrome c and SEC-SAXS data sets for the small GTPase Arf6 and the Arf GTPase exchange factors Grp1 and cytohesin-1, SVD-LC consistently provided higher quality reconstruction of protein scattering than either direct or Guinier-optimized buffer subtraction. These methods have been implemented in the context of a Python-extensible Mac OS X application known as Data Evaluation and Likelihood Analysis (DELA), which provides convenient tools for data-set selection, beam intensity normalization, SVD, and other relevant processing and analytical procedures, as well as automated Python scripts for common SAXS analyses and Guinier-optimized reconstruction of protein scattering.
Keywords: Guinier optimization; linear combination; singular value decomposition; size-exclusion chromatography; small-angle X-ray scattering.
Figures




References
-
- Biou, V., Aizel, K., Roblin, P., Thureau, A., Jacquet, E., Hansson, S., Guibert, B., Guittet, E., van Heijenoort, C., Zeghouf, M., Perez, J. & Cherfils, J. (2010). J. Mol. Biol. 402, 696–707. - PubMed
-
- Bushnell, G. W., Louie, G. V. & Brayer, G. D. (1990). J. Mol. Biol. 214, 585–595. - PubMed
-
- David, G. & Pérez, J. (2009). J. Appl. Cryst. 42, 892–900.
Grants and funding
LinkOut - more resources
Full Text Sources
