Systems medicine of inflammaging
- PMID: 26307062
- PMCID: PMC4870395
- DOI: 10.1093/bib/bbv062
Systems medicine of inflammaging
Abstract
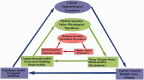
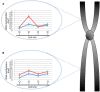
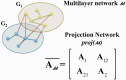
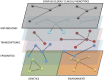
Systems Medicine (SM) can be defined as an extension of Systems Biology (SB) to Clinical-Epidemiological disciplines through a shifting paradigm, starting from a cellular, toward a patient centered framework. According to this vision, the three pillars of SM are Biomedical hypotheses, experimental data, mainly achieved by Omics technologies and tailored computational, statistical and modeling tools. The three SM pillars are highly interconnected, and their balancing is crucial. Despite the great technological progresses producing huge amount of data (Big Data) and impressive computational facilities, the Bio-Medical hypotheses are still of primary importance. A paradigmatic example of unifying Bio-Medical theory is the concept of Inflammaging. This complex phenotype is involved in a large number of pathologies and patho-physiological processes such as aging, age-related diseases and cancer, all sharing a common inflammatory pathogenesis. This Biomedical hypothesis can be mapped into an ecological perspective capable to describe by quantitative and predictive models some experimentally observed features, such as microenvironment, niche partitioning and phenotype propagation. In this article we show how this idea can be supported by computational methods useful to successfully integrate, analyze and model large data sets, combining cross-sectional and longitudinal information on clinical, environmental and omics data of healthy subjects and patients to provide new multidimensional biomarkers capable of distinguishing between different pathological conditions, e.g. healthy versus unhealthy state, physiological versus pathological aging.
Keywords: ecological model; inflammation; multi-scale; multilayer networks; networks; propagation.
© The Author 2015. Published by Oxford University Press.
Figures



References
-
- Zhang H, Gustafsson M, Nestor C, et al. Targeted omics and systems medicine: personalising care. Lancet Respir Med 2014;2:785–7. - PubMed
-
- Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol 2012;29:613–24. - PubMed
-
- Franceschi C, Bonafè M, Valensin S, et al. Inflamm-aging: an evolutionary perspective on immunosenescence. Ann NY Acad Sci 2006;908:244–54. - PubMed
-
- Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci 2014;69 (Suppl 1):S4–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

