Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor
- PMID: 26307764
- PMCID: PMC4553789
- DOI: 10.1073/pnas.1423228112
Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor
Abstract
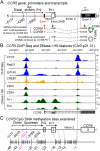
T-cell expression levels of CC chemokine receptor 5 (CCR5) are a critical determinant of HIV/AIDS susceptibility, and manifest wide variations (i) between T-cell subsets and among individuals and (ii) in T-cell activation-induced increases in expression levels. We demonstrate that a unifying mechanism for this variation is differences in constitutive and T-cell activation-induced DNA methylation status of CCR5 cis-regulatory regions (cis-regions). Commencing at an evolutionarily conserved CpG (CpG -41), CCR5 cis-regions manifest lower vs. higher methylation in T cells with higher vs. lower CCR5 levels (memory vs. naïve T cells) and in memory T cells with higher vs. lower CCR5 levels. HIV-related and in vitro induced T-cell activation is associated with demethylation of these cis-regions. CCR5 haplotypes associated with increased vs. decreased gene/surface expression levels and HIV/AIDS susceptibility magnify vs. dampen T-cell activation-associated demethylation. Methylation status of CCR5 intron 2 explains a larger proportion of the variation in CCR5 levels than genotype or T-cell activation. The ancestral, protective CCR5-HHA haplotype bears a polymorphism at CpG -41 that is (i) specific to southern Africa, (ii) abrogates binding of the transcription factor CREB1 to this cis-region, and (iii) exhibits a trend for overrepresentation in persons with reduced susceptibility to HIV and disease progression. Genotypes lacking the CCR5-Δ32 mutation but with hypermethylated cis-regions have CCR5 levels similar to genotypes heterozygous for CCR5-Δ32. In HIV-infected individuals, CCR5 cis-regions remain demethylated, despite restoration of CD4+ counts (≥800 cells per mm(3)) with antiretroviral therapy. Thus, methylation content of CCR5 cis-regions is a central epigenetic determinant of T-cell CCR5 levels, and possibly HIV-related outcomes.
Keywords: CCR5; HIV; T-cell activation; methylation; polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


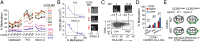




References
-
- Corbeau P, Reynes J. CCR5 antagonism in HIV infection: Ways, effects, and side effects. AIDS. 2009;23(15):1931–1943. - PubMed
-
- Paxton WA, et al. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: Association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244(1):66–73. - PubMed
-
- Reynes J, Baillat V, Portales P, Clot J, Corbeau P. Low CD4+ T-cell surface CCR5 density as a cause of resistance to in vivo HIV-1 infection. J Acquir Immune Defic Syndr. 2003;34(1):114–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI046326/AI/NIAID NIH HHS/United States
- R01 AI043279/AI/NIAID NIH HHS/United States
- P01 AI057127/AI/NIAID NIH HHS/United States
- UL1RR025767/RR/NCRR NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- UL1 RR024131/RR/NCRR NIH HHS/United States
- R01-AI043279/AI/NIAID NIH HHS/United States
- R01HL090312/HL/NHLBI NIH HHS/United States
- I01 BX000975/BX/BLRD VA/United States
- P51 OD011133/OD/NIH HHS/United States
- UL1 TR001120/TR/NCATS NIH HHS/United States
- IP1 CX000875/CX/CSRD VA/United States
- U19 AI 51794/AI/NIAID NIH HHS/United States
- R37-AI046326/AI/NIAID NIH HHS/United States
- NIH-NCI P30CA54174/CA/NCI NIH HHS/United States
- P30 MH62246/MH/NIMH NIH HHS/United States
- NIH P01AI057127/AI/NIAID NIH HHS/United States
- P30 AI027742/AI/NIAID NIH HHS/United States
- P30AI027742/AI/NIAID NIH HHS/United States
- R24 AI067039/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

