GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport
- PMID: 26308899
- PMCID: PMC4631399
- DOI: 10.1038/nature14974
GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport
Abstract
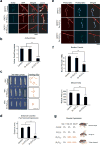
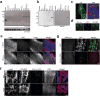
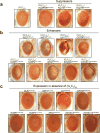
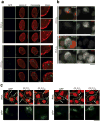
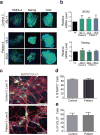
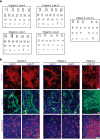
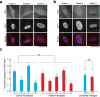
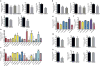
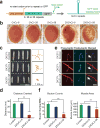
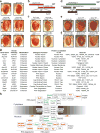
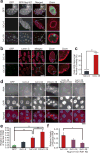
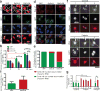
The GGGGCC (G4C2) repeat expansion in a noncoding region of C9orf72 is the most common cause of sporadic and familial forms of amyotrophic lateral sclerosis and frontotemporal dementia. The basis for pathogenesis is unknown. To elucidate the consequences of G4C2 repeat expansion in a tractable genetic system, we generated transgenic fly lines expressing 8, 28 or 58 G4C2-repeat-containing transcripts that do not have a translation start site (AUG) but contain an open-reading frame for green fluorescent protein to detect repeat-associated non-AUG (RAN) translation. We show that these transgenic animals display dosage-dependent, repeat-length-dependent degeneration in neuronal tissues and RAN translation of dipeptide repeat (DPR) proteins, as observed in patients with C9orf72-related disease. This model was used in a large-scale, unbiased genetic screen, ultimately leading to the identification of 18 genetic modifiers that encode components of the nuclear pore complex (NPC), as well as the machinery that coordinates the export of nuclear RNA and the import of nuclear proteins. Consistent with these results, we found morphological abnormalities in the architecture of the nuclear envelope in cells expressing expanded G4C2 repeats in vitro and in vivo. Moreover, we identified a substantial defect in RNA export resulting in retention of RNA in the nuclei of Drosophila cells expressing expanded G4C2 repeats and also in mammalian cells, including aged induced pluripotent stem-cell-derived neurons from patients with C9orf72-related disease. These studies show that a primary consequence of G4C2 repeat expansion is the compromise of nucleocytoplasmic transport through the nuclear pore, revealing a novel mechanism of neurodegeneration.
Figures


Comment in
-
Neurodegeneration: Problems at the nuclear pore.Nature. 2015 Sep 3;525(7567):36-7. doi: 10.1038/nature15208. Epub 2015 Aug 26. Nature. 2015. PMID: 26308896 No abstract available.
-
Neurodegenerative disease: C9orf72 repeats compromise nucleocytoplasmic transport.Nat Rev Neurol. 2015 Dec;11(12):670-2. doi: 10.1038/nrneurol.2015.219. Epub 2015 Nov 3. Nat Rev Neurol. 2015. PMID: 26526532 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

