Experimental testing of a new integrated model of the budding yeast Start transition
- PMID: 26310445
- PMCID: PMC4710230
- DOI: 10.1091/mbc.E15-06-0358
Experimental testing of a new integrated model of the budding yeast Start transition
Abstract
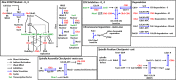
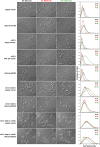
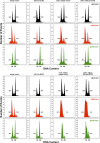
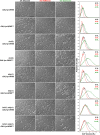
The cell cycle is composed of bistable molecular switches that govern the transitions between gap phases (G1 and G2) and the phases in which DNA is replicated (S) and partitioned between daughter cells (M). Many molecular details of the budding yeast G1-S transition (Start) have been elucidated in recent years, especially with regard to its switch-like behavior due to positive feedback mechanisms. These results led us to reevaluate and expand a previous mathematical model of the yeast cell cycle. The new model incorporates Whi3 inhibition of Cln3 activity, Whi5 inhibition of SBF and MBF transcription factors, and feedback inhibition of Whi5 by G1-S cyclins. We tested the accuracy of the model by simulating various mutants not described in the literature. We then constructed these novel mutant strains and compared their observed phenotypes to the model's simulations. The experimental results reported here led to further changes of the model, which will be fully described in a later article. Our study demonstrates the advantages of combining model design, simulation, and testing in a coordinated effort to better understand a complex biological network.
© 2015 Adames et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures


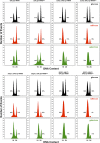
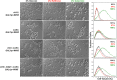
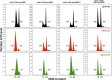
References
-
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

