Deep proteomic profiling of vasopressin-sensitive collecting duct cells. II. Bioinformatic analysis of vasopressin signaling
- PMID: 26310817
- PMCID: PMC4683213
- DOI: 10.1152/ajpcell.00214.2015
Deep proteomic profiling of vasopressin-sensitive collecting duct cells. II. Bioinformatic analysis of vasopressin signaling
Abstract
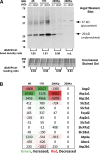
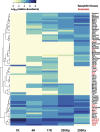
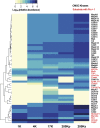
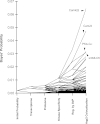
Vasopressin controls osmotic water transport in the renal collecting duct through regulation of aquaporin-2 (AQP2). We carried out bioinformatic analysis of quantitative proteomic data from the accompanying article to investigate the mechanisms involved. The experiments used stable isotope labeling by amino acids in cell culture in cultured mpkCCD cells to quantify each protein species in each of five differential-centrifugation (DC) fractions with or without the vasopressin analog 1-desamino-8-d-arginine-vasopressin (dDAVP). The mass spectrometry data and parallel Western blot experiments confirmed that dDAVP addition is associated with an increase in AQP2 abundance in the 17,000-g pellet and a corresponding decrease in the 200,000-g pellet. Remarkably, all subunits of the cytoplasmic ribosome also increased in the 17,000-g pellet in response to dDAVP (P < 10(-34)), with a concomitant decrease in the 200,000-g pellet. Eukaryotic translation initiation complex 3 (eIF3) subunits underwent parallel changes (P < 10(-6)). These findings are consistent with translocation of assembled ribosomes and eIF3 complexes into the rough endoplasmic reticulum in response to dDAVP. Conversely, there was a systematic decrease in small GTPase abundances in the 17,000-g fraction. In contrast, most proteins, including protein kinases, showed no systematic redistribution among DC fractions. Of the 521 protein kinases coded by the mouse genome, 246 were identified, but many fewer were found to colocalize with AQP2 among DC fractions. Bayes' rule was used to integrate the new colocalization data with prior data to identify protein kinases most likely to phosphorylate aquaporin-2 at Ser(256) (Camk2b > Camk2d > Prkaca) and Ser(261) (Mapk1 = Mapk3 > Mapk14).
Keywords: aquaporin-2; mass spectrometry; phosphorylation; protein kinase; ribosome; small GTPase; translation.
Figures








Comment in
-
Water transport running deep. Focus on "Deep proteomic profiling of vasopressin-sensitive collecting duct cells".Am J Physiol Cell Physiol. 2015 Dec 15;309(12):C783-4. doi: 10.1152/ajpcell.00280.2015. Epub 2015 Sep 30. Am J Physiol Cell Physiol. 2015. PMID: 26423579 No abstract available.
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science, 2002.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

