MLL leukemia induction by genome editing of human CD34+ hematopoietic cells
- PMID: 26311362
- PMCID: PMC4591792
- DOI: 10.1182/blood-2015-05-646398
MLL leukemia induction by genome editing of human CD34+ hematopoietic cells
Abstract
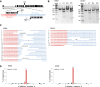
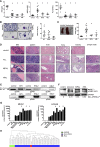
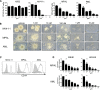
Chromosomal rearrangements involving the mixed-lineage leukemia (MLL) gene occur in primary and treatment-related leukemias and confer a poor prognosis. Studies based primarily on mouse models have substantially advanced our understanding of MLL leukemia pathogenesis, but often use supraphysiological oncogene expression with uncertain implications for human leukemia. Genome editing using site-specific nucleases provides a powerful new technology for gene modification to potentially model human disease, however, this approach has not been used to re-create acute leukemia in human cells of origin comparable to disease observed in patients. We applied transcription activator-like effector nuclease-mediated genome editing to generate endogenous MLL-AF9 and MLL-ENL oncogenes through insertional mutagenesis in primary human hematopoietic stem and progenitor cells (HSPCs) derived from human umbilical cord blood. Engineered HSPCs displayed altered in vitro growth potentials and induced acute leukemias following transplantation in immunocompromised mice at a mean latency of 16 weeks. The leukemias displayed phenotypic and morphologic similarities with patient leukemia blasts including a subset with mixed phenotype, a distinctive feature seen in clinical disease. The leukemic blasts expressed an MLL-associated transcriptional program with elevated levels of crucial MLL target genes, displayed heightened sensitivity to DOT1L inhibition, and demonstrated increased oncogenic potential ex vivo and in secondary transplant assays. Thus, genome editing to create endogenous MLL oncogenes in primary human HSPCs faithfully models acute MLL-rearranged leukemia and provides an experimental platform for prospective studies of leukemia initiation and stem cell biology in a genetic subtype of poor prognosis leukemia.
© 2015 by The American Society of Hematology.
Figures



References
-
- Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18(9):965–974. - PubMed
-
- Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106(3):614–626. - PubMed
-
- De Braekeleer M, Morel F, Le Bris MJ, Herry A, Douet-Guilbert N. The MLL gene and translocations involving chromosomal band 11q23 in acute leukemia. Anticancer Res. 2005;25(3B):1931–1944. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

