Inactivation of the Human Cytomegalovirus US20 Gene Hampers Productive Viral Replication in Endothelial Cells
- PMID: 26311874
- PMCID: PMC4621124
- DOI: 10.1128/JVI.01141-15
Inactivation of the Human Cytomegalovirus US20 Gene Hampers Productive Viral Replication in Endothelial Cells
Abstract
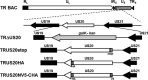
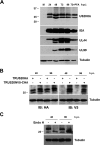
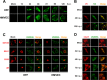
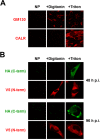
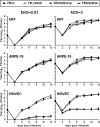
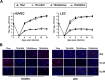
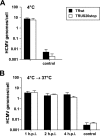
The human cytomegalovirus (HCMV) US12 gene family includes a group of 10 contiguous genes (US12 to US21) encoding predicted seven-transmembrane-domain (7TMD) proteins that are nonessential for replication within cultured fibroblasts. Nevertheless, inactivation of some US12 family members affects virus replication in other cell types; e.g., deletion of US16 or US18 abrogates virus growth in endothelial and epithelial cells or in human gingival tissue, respectively, suggesting a role for some US12 proteins in HCMV cell tropism. Here, we provide evidence that another member, US20, impacts the ability of a clinical strain of HCMV to replicate in endothelial cells. Through the use of recombinant HCMV encoding tagged versions of the US20 protein, we investigated the expression pattern, localization, and topology of the US20-encoded protein (pUS20). We show that pUS20 is expressed as a partially glycosylated 7TMD protein which accumulates late in infection in endoplasmic reticulum-derived peripheral structures localized outside the cytoplasmic virus assembly compartment (cVAC). US20-deficient mutants generated in the TR clinical strain of HCMV exhibited major growth defects in different types of endothelial cells, whereas they replicated normally in fibroblasts and epithelial cells. While the attachment and entry phases in endothelial cells were not significantly affected by the absence of US20 protein, US20-null viruses failed to replicate viral DNA and express representative E and L mRNAs and proteins. Taken together, these results indicate that US20 sustains the HCMV replication cycle at a stage subsequent to entry but prior to E gene expression and viral DNA synthesis in endothelial cells.
Importance: Human cytomegalovirus (HCMV) is a major pathogen in newborns and immunocompromised individuals. A hallmark of HCMV pathogenesis is its ability to productively replicate in an exceptionally broad range of target cells, including endothelial cells, which represent a key target for viral dissemination and replication in the host, and to contribute to both viral persistence and associated inflammation and vascular diseases. Replication in endothelial cells depends on the activities of a set of viral proteins that regulate different stages of the HCMV replication cycle in an endothelial cell type-specific manner and thereby act as determinants of viral tropism. Here, we report the requirement of a HCMV protein as a postentry tropism factor in endothelial cells. The identification and characterization of HCMV endotheliotropism-regulating proteins will advance our understanding of the molecular mechanisms of HCMV-related pathogenesis and help lead to the design of new antiviral strategies able to exploit these functions.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. - PubMed
-
- Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Murphy E, Shenk T. 2008. Human cytomegalovirus genomes. Curr Top Microbiol Immunol 325:1–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

