Reorganization of Nuclear Pore Complexes and the Lamina in Late-Stage Parvovirus Infection
- PMID: 26311881
- PMCID: PMC4645667
- DOI: 10.1128/JVI.01608-15
Reorganization of Nuclear Pore Complexes and the Lamina in Late-Stage Parvovirus Infection
Abstract
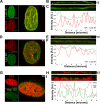
Canine parvovirus (CPV) infection induces reorganization of nuclear structures. Our studies indicated that late-stage infection induces accumulation of nuclear pore complexes (NPCs) and lamin B1 concomitantly with a decrease of lamin A/C levels on the apical side of the nucleus. Newly formed CPV capsids are located in close proximity to NPCs on the apical side. These results suggest that parvoviruses cause apical enrichment of NPCs and reorganization of nuclear lamina, presumably to facilitate the late-stage infection.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Nuclear pore protein TPR associates with lamin B1 and affects nuclear lamina organization and nuclear pore distribution.Cell Mol Life Sci. 2019 Jun;76(11):2199-2216. doi: 10.1007/s00018-019-03037-0. Epub 2019 Feb 14. Cell Mol Life Sci. 2019. PMID: 30762072 Free PMC article.
-
Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins.J Cell Biol. 2021 Apr 5;220(4):e202007082. doi: 10.1083/jcb.202007082. J Cell Biol. 2021. PMID: 33570570 Free PMC article.
-
Analysis of the VP2 protein gene of canine parvovirus strains from affected dogs in Japan.Res Vet Sci. 2013 Apr;94(2):368-71. doi: 10.1016/j.rvsc.2012.09.013. Epub 2012 Oct 12. Res Vet Sci. 2013. PMID: 23063259
-
Parvovirus infection in domestic companion animals.Vet Clin North Am Small Anim Pract. 2008 Jul;38(4):837-50, viii-ix. doi: 10.1016/j.cvsm.2008.03.008. Vet Clin North Am Small Anim Pract. 2008. PMID: 18501282 Review.
-
The nuclear envelopathies and human diseases.J Biomed Sci. 2009 Oct 22;16(1):96. doi: 10.1186/1423-0127-16-96. J Biomed Sci. 2009. PMID: 19849840 Free PMC article. Review.
Cited by
-
Structural and Mechanical Aberrations of the Nuclear Lamina in Disease.Cells. 2020 Aug 11;9(8):1884. doi: 10.3390/cells9081884. Cells. 2020. PMID: 32796718 Free PMC article. Review.
-
Construction of Nuclear Envelope Shape by a High-Genus Vesicle with Pore-Size Constraint.Biophys J. 2016 Aug 23;111(4):824-831. doi: 10.1016/j.bpj.2016.07.010. Biophys J. 2016. PMID: 27558725 Free PMC article.
-
Nuclear Cytoskeleton in Virus Infection.Int J Mol Sci. 2022 Jan 5;23(1):578. doi: 10.3390/ijms23010578. Int J Mol Sci. 2022. PMID: 35009004 Free PMC article. Review.
-
Cytoplasmic Parvovirus Capsids Recruit Importin Beta for Nuclear Delivery.J Virol. 2020 Jan 31;94(4):e01532-19. doi: 10.1128/JVI.01532-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31748386 Free PMC article.
-
Nuclear envelope budding and its cellular functions.Nucleus. 2023 Dec;14(1):2178184. doi: 10.1080/19491034.2023.2178184. Nucleus. 2023. PMID: 36814098 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

