Viral Polymerase-Helicase Complexes Regulate Replication Fidelity To Overcome Intracellular Nucleotide Depletion
- PMID: 26311883
- PMCID: PMC4645662
- DOI: 10.1128/JVI.01553-15
Viral Polymerase-Helicase Complexes Regulate Replication Fidelity To Overcome Intracellular Nucleotide Depletion
Abstract
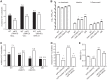
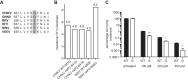
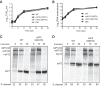
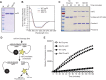
To date, the majority of work on RNA virus replication fidelity has focused on the viral RNA polymerase, while the potential role of other viral replicase proteins in this process is poorly understood. Previous studies used resistance to broad-spectrum RNA mutagens, such as ribavirin, to identify polymerases with increased fidelity that avoid misincorporation of such base analogues. We identified a novel variant in the alphavirus viral helicase/protease, nonstructural protein 2 (nsP2) that operates in concert with the viral polymerase nsP4 to further alter replication complex fidelity, a functional linkage that was conserved among the alphavirus genus. Purified chikungunya virus nsP2 presented delayed helicase activity of the high-fidelity enzyme, and yet purified replication complexes manifested stronger RNA polymerization kinetics. Because mutagenic nucleoside analogs such as ribavirin also affect intracellular nucleotide pools, we addressed the link between nucleotide depletion and replication fidelity by using purine and pyrimidine biosynthesis inhibitors. High-fidelity viruses were more resistant to these conditions, and viral growth could be rescued by the addition of exogenous nucleosides, suggesting that mutagenesis by base analogues requires nucleotide pool depletion. This study describes a novel function for nsP2, highlighting the role of other components of the replication complex in regulating viral replication fidelity, and suggests that viruses can alter their replication complex fidelity to overcome intracellular nucleotide-depleting conditions.
Importance: Previous studies using the RNA mutagen ribavirin to select for drug-resistant variants have highlighted the essential role of the viral RNA-dependent RNA polymerase in regulating replication fidelity. However, the role of other viral replicase components in replication fidelity has not been studied in detail. We identified here an RNA mutagen-resistant variant of the nsP2 helicase/protease that conferred increased fidelity and yet could not operate in the same manner as high-fidelity polymerases. We show that the alphavirus helicase is a key component of the fidelity-regulating machinery. Our data show that the RNA mutagenic activity of compounds such as ribavirin is coupled to and potentiated by nucleotide depletion and that RNA viruses can fine-tune their replication fidelity when faced with an intracellular environment depleted of nucleotides.
Copyright © 2015, Stapleford et al.
Figures



References
-
- Arias A, Arnold JJ, Sierra M, Smidansky ED, Domingo E, Cameron CE. 2008. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J Virol 82:12346–12355. doi: 10.1128/JVI.01297-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

