Oligomerization of Mumps Virus Phosphoprotein
- PMID: 26311887
- PMCID: PMC4621118
- DOI: 10.1128/JVI.01719-15
Oligomerization of Mumps Virus Phosphoprotein
Abstract
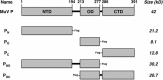
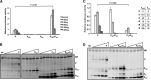
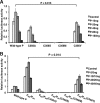
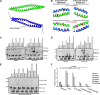
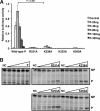
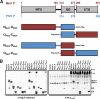
The mumps virus (MuV) genome encodes a phosphoprotein (P) that is important for viral RNA synthesis. P forms the viral RNA-dependent RNA polymerase with the large protein (L). P also interacts with the viral nucleoprotein (NP) and self-associates to form a homotetramer. The P protein consists of three domains, the N-terminal domain (P(N)), the oligomerization domain (P(O)), and the C-terminal domain (P(C)). While P(N) is known to relax the NP-bound RNA genome, the roles of P(O) and P(C) are not clear. In this study, we investigated the roles of P(O) and P(C) in viral RNA synthesis using mutational analysis and a minigenome system. We found that P(N) and P(C) functions can be trans-complemented. However, this complementation requires P(O), indicating that P(O) is essential for P function. Using this trans-complementation system, we found that P forms parallel dimers (P(N) to P(N) and P(C) to P(C)). Furthermore, we found that residues R231, K238, K253, and K260 in P(O) are critical for P's functions. We identified P(C) to be the domain that interacts with L. These results provide structure-function insights into the role of MuV P.
Importance: MuV, a paramyxovirus, is an important human pathogen. The P protein of MuV is critical for viral RNA synthesis. In this work, we established a novel minigenome system that allows the domains of P to be complemented in trans. Using this system, we confirmed that MuV P forms parallel dimers. An understanding of viral RNA synthesis will allow the design of better vaccines and the development of antivirals.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication.J Virol. 2014 Apr;88(8):4414-22. doi: 10.1128/JVI.03673-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501400 Free PMC article.
-
Regulation of Viral RNA Synthesis by the V Protein of Parainfluenza Virus 5.J Virol. 2015 Dec;89(23):11845-57. doi: 10.1128/JVI.01832-15. Epub 2015 Sep 16. J Virol. 2015. PMID: 26378167 Free PMC article.
-
Roles of Phosphorylation of the Nucleocapsid Protein of Mumps Virus in Regulating Viral RNA Transcription and Replication.J Virol. 2015 Jul;89(14):7338-47. doi: 10.1128/JVI.00686-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948749 Free PMC article.
-
Regulation of Mumps Virus Replication and Transcription by Kinase RPS6KB1.J Virol. 2020 Jun 1;94(12):e00387-20. doi: 10.1128/JVI.00387-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32295907 Free PMC article.
-
Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression.Future Microbiol. 2010 Jan;5(1):9-13. doi: 10.2217/fmb.09.93. Future Microbiol. 2010. PMID: 20020826 Free PMC article. Review.
Cited by
-
Crystal Structure of the Marburg Virus VP35 Oligomerization Domain.J Virol. 2017 Jan 3;91(2):e01085-16. doi: 10.1128/JVI.01085-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847355 Free PMC article.
-
Structures of the mumps virus polymerase complex via cryo-electron microscopy.Nat Commun. 2024 May 17;15(1):4189. doi: 10.1038/s41467-024-48389-9. Nat Commun. 2024. PMID: 38760379 Free PMC article.
-
Regulation of measles virus gene expression by P protein coiled-coil properties.Sci Adv. 2019 May 8;5(5):eaaw3702. doi: 10.1126/sciadv.aaw3702. eCollection 2019 May. Sci Adv. 2019. PMID: 31086822 Free PMC article.
-
Human Parainfluenza Virus 3 Phosphoprotein Is a Tetramer and Shares Structural and Interaction Features with Ebola Phosphoprotein VP35.Biomolecules. 2021 Oct 29;11(11):1603. doi: 10.3390/biom11111603. Biomolecules. 2021. PMID: 34827601 Free PMC article.
-
The Nucleoprotein and Phosphoprotein of Measles Virus.Front Microbiol. 2019 Aug 21;10:1832. doi: 10.3389/fmicb.2019.01832. eCollection 2019. Front Microbiol. 2019. PMID: 31496998 Free PMC article. Review.
References
-
- Carbone KM, Wolinsky JS. 2001. Mumps virus, p 1381–1441. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Lamb RA, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication, p 1305–1340. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

