Proteomic analysis of quail calcified eggshell matrix: a comparison to chicken and turkey eggshell proteomes
- PMID: 26312056
- PMCID: PMC4550075
- DOI: 10.1186/s12953-015-0078-1
Proteomic analysis of quail calcified eggshell matrix: a comparison to chicken and turkey eggshell proteomes
Abstract
Background: Eggshell mineralization in commercially important species such as chicken, turkey or quail is of interest as a general model of calcium carbonate biomineralization. Knowledge of proteins and molecular mechanisms in eggshell assembly may also pave the way to manipulation of thickness of the calcified layer or other features. Comparison of eggshell matrix proteomes of different species may contribute to a better understanding of the mineralization process. The recent publication of the quail genome sequence now enables the proteomic analysis of the quail shell matrix and this comparison with those of chicken and turkey.
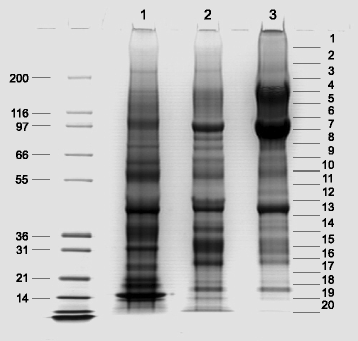
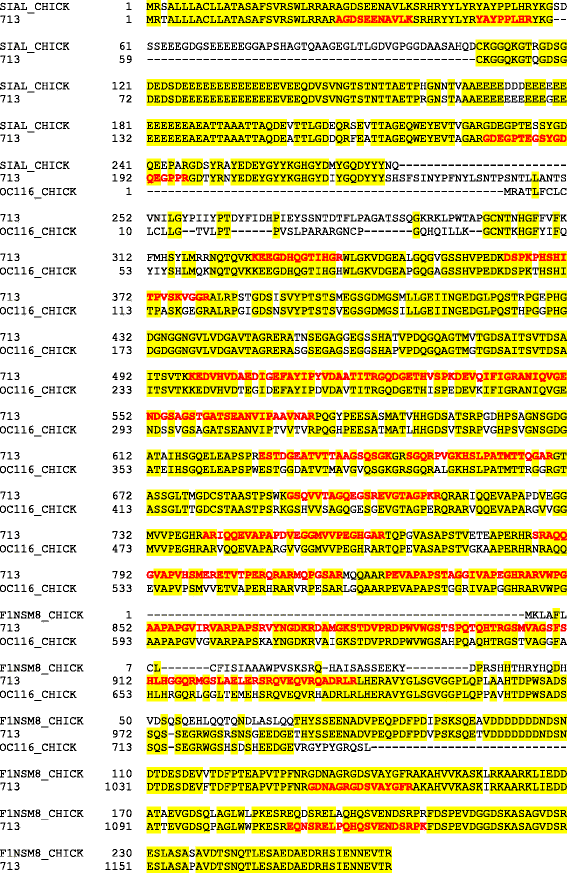
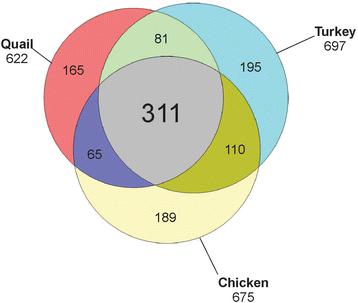
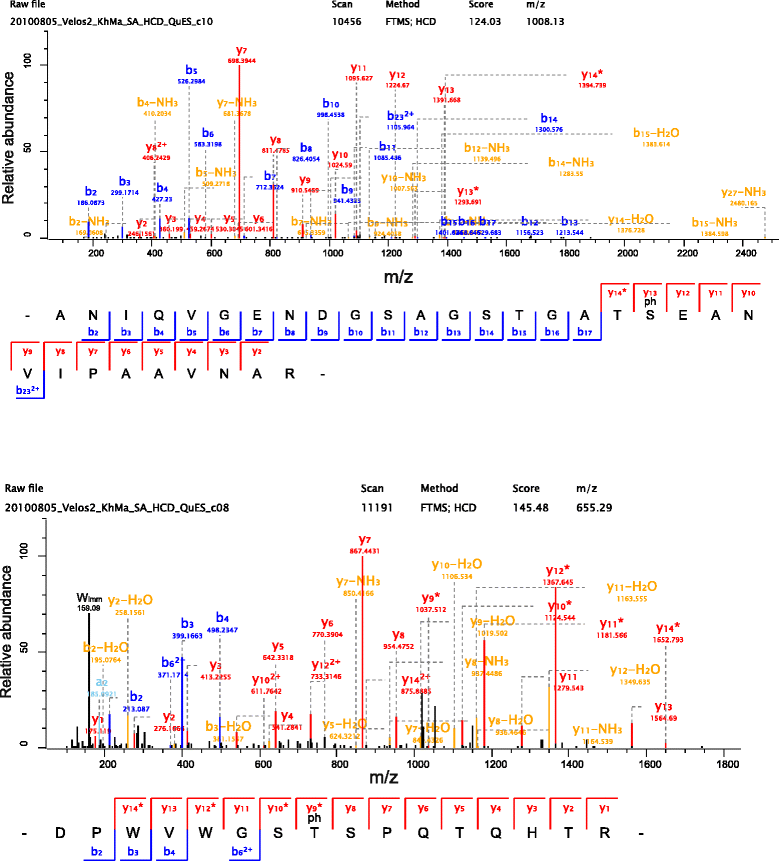
Results: The quail eggshell proteome comprised 622 identified proteins, 311 of which were shared with chicken and turkey eggshell proteomes. Forty-eight major proteins (iBAQ-derived abundance higher than 0.1 % of total identified proteome) together covered 94 % of total proteome mass. Fifteen of these are also among the most abundant proteins in chicken and turkey eggshell matrix. Only three proteins with a percentage higher than 1.0 % of the total had not previously been identified as eggshell matrix proteins. These were an uncharacterized member of the latexin family, an uncharacterized protease inhibitor containing a Kunitz domain, and gastric intrinsic factor. The most abundant proteins were ovocleidin-116, ovalbumin and ovocalyxin-36 representing approximately 31, 13 and 8 % of the total identified proteome, respectively. The major phosphoproteins were ovocleidin-116 and osteopontin. While osteopontin phosphorylation sites were predominantly conserved between chicken and quail sequences, conservation was less in ovocleidin-116.
Conclusions: Ovocleidin-116 and ovocalyxin-36 are among the most abundant eggshell matrix proteins in all three species of the family Phasianidae analyzed so far, indicating that their presently unknown function is essential for eggshell mineralization. Evidence for other chicken eggshell-specific proteins in quail was inconclusive. Therefore measurement of additional eggshell proteomes, especially from species of different families and preferentially from outside the order Galliformes, will be necessary.
Figures





Similar articles
-
The calcified eggshell matrix proteome of a songbird, the zebra finch (Taeniopygia guttata).Proteome Sci. 2015 Dec 1;13:29. doi: 10.1186/s12953-015-0086-1. eCollection 2015. Proteome Sci. 2015. PMID: 26628892 Free PMC article.
-
The proteome of the calcified layer organic matrix of turkey (Meleagris gallopavo) eggshell.Proteome Sci. 2013 Aug 27;11(1):40. doi: 10.1186/1477-5956-11-40. Proteome Sci. 2013. PMID: 23981693 Free PMC article.
-
Phosphoproteins of the chicken eggshell calcified layer.Proteomics. 2007 Jan;7(1):106-15. doi: 10.1002/pmic.200600635. Proteomics. 2007. PMID: 17152097
-
Antimicrobial Proteins and Peptides in Avian Eggshell: Structural Diversity and Potential Roles in Biomineralization.Front Immunol. 2022 Jul 27;13:946428. doi: 10.3389/fimmu.2022.946428. eCollection 2022. Front Immunol. 2022. PMID: 35967448 Free PMC article. Review.
-
The eggshell: structure, composition and mineralization.Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1266-80. doi: 10.2741/3985. Front Biosci (Landmark Ed). 2012. PMID: 22201802 Review.
Cited by
-
Protein sequences bound to mineral surfaces persist into deep time.Elife. 2016 Sep 27;5:e17092. doi: 10.7554/eLife.17092. Elife. 2016. PMID: 27668515 Free PMC article.
-
The calcified eggshell matrix proteome of a songbird, the zebra finch (Taeniopygia guttata).Proteome Sci. 2015 Dec 1;13:29. doi: 10.1186/s12953-015-0086-1. eCollection 2015. Proteome Sci. 2015. PMID: 26628892 Free PMC article.
-
Copy Number Variants in Four Italian Turkey Breeds.Animals (Basel). 2021 Feb 3;11(2):391. doi: 10.3390/ani11020391. Animals (Basel). 2021. PMID: 33546454 Free PMC article.
-
The glycoproteins EDIL3 and MFGE8 regulate vesicle-mediated eggshell calcification in a new model for avian biomineralization.J Biol Chem. 2019 Oct 4;294(40):14526-14545. doi: 10.1074/jbc.RA119.009799. Epub 2019 Jul 29. J Biol Chem. 2019. PMID: 31358619 Free PMC article.
-
Avian eggshell biomineralization: an update on its structure, mineralogy and protein tool kit.BMC Mol Cell Biol. 2021 Feb 12;22(1):11. doi: 10.1186/s12860-021-00350-0. BMC Mol Cell Biol. 2021. PMID: 33579194 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

