Candida albicans Inhibits Pseudomonas aeruginosa Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- PMID: 26313907
- PMCID: PMC4552174
- DOI: 10.1371/journal.ppat.1005129
Candida albicans Inhibits Pseudomonas aeruginosa Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
Abstract
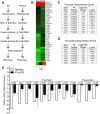
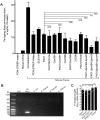
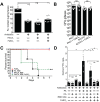
Bacterial-fungal interactions have important physiologic and medical ramifications, but the mechanisms of these interactions are poorly understood. The gut is host to trillions of microorganisms, and bacterial-fungal interactions are likely to be important. Using a neutropenic mouse model of microbial gastrointestinal colonization and dissemination, we show that the fungus Candida albicans inhibits the virulence of the bacterium Pseudomonas aeruginosa by inhibiting P. aeruginosa pyochelin and pyoverdine gene expression, which plays a critical role in iron acquisition and virulence. Accordingly, deletion of both P. aeruginosa pyochelin and pyoverdine genes attenuates P. aeruginosa virulence. Heat-killed C. albicans has no effect on P. aeruginosa, whereas C. albicans secreted proteins directly suppress P. aeruginosa pyoverdine and pyochelin expression and inhibit P. aeruginosa virulence in mice. Interestingly, suppression or deletion of pyochelin and pyoverdine genes has no effect on P. aeruginosa's ability to colonize the GI tract but does decrease P. aeruginosa's cytotoxic effect on cultured colonocytes. Finally, oral iron supplementation restores P. aeruginosa virulence in P. aeruginosa and C. albicans colonized mice. Together, our findings provide insight into how a bacterial-fungal interaction can modulate bacterial virulence in the intestine. Previously described bacterial-fungal antagonistic interactions have focused on growth inhibition or colonization inhibition/modulation, yet here we describe a novel observation of fungal-inhibition of bacterial effectors critical for virulence but not important for colonization. These findings validate the use of a mammalian model system to explore the complexities of polymicrobial, polykingdom infections in order to identify new therapeutic targets for preventing microbial disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
The complexities of bacterial-fungal interactions in the mammalian gastrointestinal tract.Microb Cell. 2016 Mar 16;3(5):191-195. doi: 10.15698/mic2016.05.497. Microb Cell. 2016. PMID: 28357354 Free PMC article. No abstract available.
References
-
- Hermann C, Hermann J, Munzel U, Ruchel R (1999) Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42: 619–627. - PubMed
-
- Azoulay E, Timsit JF, Tafflet M, de Lassence A, Darmon M, et al. (2006) Candida colonization of the respiratory tract and subsequent pseudomonas ventilator-associated pneumonia. Chest 129: 110–117. - PubMed
-
- Bauernfeind A, Bertele RM, Harms K, Horl G, Jungwirth R, et al. (1987) Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15: 270–277. - PubMed
-
- Gupta N, Haque A, Mukhopadhyay G, Narayan RP, Prasad R (2005) Interactions between bacteria and Candida in the burn wound. Burns 31: 375–378. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

