Bioinformatics Analysis of the Complete Genome Sequence of the Mango Tree Pathogen Pseudomonas syringae pv. syringae UMAF0158 Reveals Traits Relevant to Virulence and Epiphytic Lifestyle
- PMID: 26313942
- PMCID: PMC4551802
- DOI: 10.1371/journal.pone.0136101
Bioinformatics Analysis of the Complete Genome Sequence of the Mango Tree Pathogen Pseudomonas syringae pv. syringae UMAF0158 Reveals Traits Relevant to Virulence and Epiphytic Lifestyle
Abstract
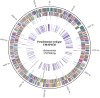
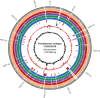
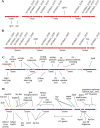
The genome sequence of more than 100 Pseudomonas syringae strains has been sequenced to date; however only few of them have been fully assembled, including P. syringae pv. syringae B728a. Different strains of pv. syringae cause different diseases and have different host specificities; so, UMAF0158 is a P. syringae pv. syringae strain related to B728a but instead of being a bean pathogen it causes apical necrosis of mango trees, and the two strains belong to different phylotypes of pv.syringae and clades of P. syringae. In this study we report the complete sequence and annotation of P. syringae pv. syringae UMAF0158 chromosome and plasmid pPSS158. A comparative analysis with the available sequenced genomes of other 25 P. syringae strains, both closed (the reference genomes DC3000, 1448A and B728a) and draft genomes was performed. The 5.8 Mb UMAF0158 chromosome has 59.3% GC content and comprises 5017 predicted protein-coding genes. Bioinformatics analysis revealed the presence of genes potentially implicated in the virulence and epiphytic fitness of this strain. We identified several genetic features, which are absent in B728a, that may explain the ability of UMAF0158 to colonize and infect mango trees: the mangotoxin biosynthetic operon mbo, a gene cluster for cellulose production, two different type III and two type VI secretion systems, and a particular T3SS effector repertoire. A mutant strain defective in the rhizobial-like T3SS Rhc showed no differences compared to wild-type during its interaction with host and non-host plants and worms. Here we report the first complete sequence of the chromosome of a pv. syringae strain pathogenic to a woody plant host. Our data also shed light on the genetic factors that possibly determine the pathogenic and epiphytic lifestyle of UMAF0158. This work provides the basis for further analysis on specific mechanisms that enable this strain to infect woody plants and for the functional analysis of host specificity in the P. syringae complex.
Conflict of interest statement
Figures



References
-
- Hirano SS, Upper CD. Population biology and epidemiology of Pseudomonas syringae . Annu Rev Phytopathol. 1990; 28: 155–177.
-
- Agrios GN. Plant Pathology. 5th ed. San Diego, CA, USA: Elsevier Academic Press; 2005.
-
- Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, et al. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J. Plant Pathol. 2010; 92: 551–592.
-
- Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont PAD. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int J Syst Bacteriol. 1999; 49: 469–478. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

