Lysine 206 in Arabidopsis phytochrome A is the major site for ubiquitin-dependent protein degradation
- PMID: 26314334
- PMCID: PMC4892773
- DOI: 10.1093/jb/mvv085
Lysine 206 in Arabidopsis phytochrome A is the major site for ubiquitin-dependent protein degradation
Abstract
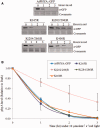
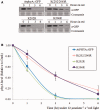
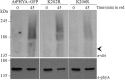
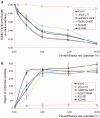
Phytochrome A (phyA) is a light labile phytochrome that mediates plant development under red/far-red light condition. Degradation of phyA is initiated by red light-induced phyA-ubiquitin conjugation through the 26S proteasome pathway. The N-terminal of phyA is known to be important in phyA degradation. To determine the specific lysine residues in the N-terminal domain of phyA involved in light-induced ubiquitination and protein degradation, we aligned the amino acid sequence of the N-terminal domain of Arabidopsis phyA with those of phyA from other plant species. Based on the alignment results, phytochrome over-expressing Arabidopsis plants were generated. In particular, wild-type and mutant (substitutions of conserved lysines by arginines) phytochromes fused with GFP were expressed in phyA(-)211 Arabidopsis plants. Degradation kinetics of over-expressed phyA proteins revealed that degradation of the K206R phyA mutant protein was delayed. Delayed phyA degradation of the K206R phyA mutant protein resulted in reduction of red-light-induced phyA-ubiquitin conjugation. Furthermore, seedlings expressing the K206R phyA mutant protein showed an enhanced phyA response under far-red light, resulting in inhibition of hypocotyl elongation as well as cotyledon opening. Together, these results suggest that lysine 206 is the main lysine for rapid ubiquitination and protein degradation of Arabidopsis phytochrome A.
Keywords: light-induced phyA degradation; phytochrome A; proteasome; ubiquitination.
© The Authors 2015. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures


References
-
- Deng X.W., Quail P.H. (1999) Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129 - PubMed
-
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010) Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66 - PubMed
-
- Neff M.M., Fankhauser C., Chory J. (2000) Light: an indicator of time and place. Genes Dev. 14, 257–271 - PubMed
-
- Franklin K.A., Larner V.S., Whitelam G.C. (2005) The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49, 653–664 - PubMed
-
- Demarsy E., Fankhauser C. (2009) Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12, 69–74 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

