Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor
- PMID: 26314708
- PMCID: PMC4573854
- DOI: 10.1101/gad.266999.115
Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor
Abstract
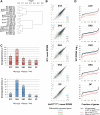
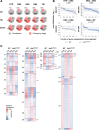
The DNA-binding protein Ikaros is a potent tumor suppressor and hematopoietic regulator. However, the mechanisms by which Ikaros functions remain poorly understood, due in part to its atypical DNA-binding properties and partnership with the poorly understood Mi-2/NuRD complex. In this study, we analyzed five sequential stages of thymocyte development in a mouse strain containing a targeted deletion of Ikaros zinc finger 4, which exhibits a select subset of abnormalities observed in Ikaros-null mice. By examining thymopoiesis in vivo and in vitro, diverse abnormalities were observed at each developmental stage. RNA sequencing revealed that each stage is characterized by the misregulation of a limited number of genes, with a strong preference for stage-specific rather than lineage-specific genes. Strikingly, individual genes rarely exhibited Ikaros dependence at all stages. Instead, a consistent feature of the aberrantly expressed genes was a reduced magnitude of expression level change during developmental transitions. These results, combined with analyses of the interplay between Ikaros loss of function and Notch signaling, suggest that Ikaros may not be a conventional activator or repressor of defined sets of genes. Instead, a primary function may be to sharpen the dynamic range of gene expression changes during developmental transitions via atypical molecular mechanisms that remain undefined.
Keywords: Ikaros; gene regulation; leukemogenesis; lymphocyte development; transcription.
© 2015 Arenzana et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Aifantis I, Mandal M, Sawai K, Ferrando A, Vilimas T. 2006. Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol Rev 209: 159–169. - PubMed
-
- Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. 1999. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10: 333–343. - PubMed
-
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845–854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
