Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions
- PMID: 26314830
- PMCID: PMC4617971
- DOI: 10.1101/gr.192294.115
Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions
Abstract
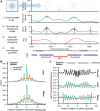
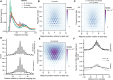
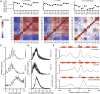
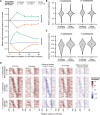
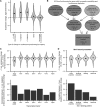
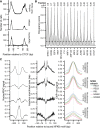
Transcription factors canonically bind nucleosome-free DNA, making the positioning of nucleosomes within regulatory regions crucial to the regulation of gene expression. Using the assay of transposase accessible chromatin (ATAC-seq), we observe a highly structured pattern of DNA fragment lengths and positions around nucleosomes in Saccharomyces cerevisiae, and use this distinctive two-dimensional nucleosomal "fingerprint" as the basis for a new nucleosome-positioning algorithm called NucleoATAC. We show that NucleoATAC can identify the rotational and translational positions of nucleosomes with up to base-pair resolution and provide quantitative measures of nucleosome occupancy in S. cerevisiae, Schizosaccharomyces pombe, and human cells. We demonstrate the application of NucleoATAC to a number of outstanding problems in chromatin biology, including analysis of sequence features underlying nucleosome positioning, promoter chromatin architecture across species, identification of transient changes in nucleosome occupancy and positioning during a dynamic cellular response, and integrated analysis of nucleosome occupancy and transcription factor binding.
© 2015 Schep et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
