Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency
- PMID: 26314899
- PMCID: PMC4551974
- DOI: 10.1038/srep13533
Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency
Abstract
The transcription factors OCT4 and SOX2 are required for generating induced pluripotent stem cells (iPSCs) and for maintaining embryonic stem cells (ESCs). OCT4 and SOX2 associate and bind to DNA in different configurations depending on the arrangement of their individual DNA binding elements. Here we have investigated the role of the different OCT4-SOX2-DNA assemblies in regulating and inducing pluripotency. To this end, we have generated SOX2 mutants that interfere with specific OCT4-SOX2 heterodimer configurations and assessed their ability to generate iPSCs and to rescue ESC self-renewal. Our results demonstrate that the OCT4-SOX2 configuration that dimerizes on a Hoxb1-like composite, a canonical element with juxtaposed individual binding sites, plays a more critical role in the induction and maintenance of pluripotency than any other OCT4-SOX2 configuration. Overall, the results of this study provide new insight into the protein interactions required to establish a de novo pluripotent network and to maintain a true pluripotent cell fate.
Figures
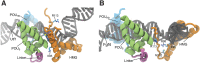
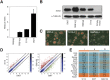

 ,
,  and
and  , respectively. Of note, full-length SOX2 binding alone cannot be observed in this assay. (C) Pairwise scatter plots exhibiting the expression level of genes co-bounded by OCT4 and SOX2 on Hoxb1-like motif (left), Utf1-like motif (center) and Fgf4-like motif (right). A red and a green line represent a 2-fold expression level increase or decrease in comparison with SWT. Genes up- and downregulated are shown by red and green dots, respectively. Expression levels below 5 were considered as background.
, respectively. Of note, full-length SOX2 binding alone cannot be observed in this assay. (C) Pairwise scatter plots exhibiting the expression level of genes co-bounded by OCT4 and SOX2 on Hoxb1-like motif (left), Utf1-like motif (center) and Fgf4-like motif (right). A red and a green line represent a 2-fold expression level increase or decrease in comparison with SWT. Genes up- and downregulated are shown by red and green dots, respectively. Expression levels below 5 were considered as background.References
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
-
- Huangfu D. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

