Whole-exome analysis reveals novel somatic genomic alterations associated with outcome in immunochemotherapy-treated diffuse large B-cell lymphoma
- PMID: 26314988
- PMCID: PMC4558593
- DOI: 10.1038/bcj.2015.69
Whole-exome analysis reveals novel somatic genomic alterations associated with outcome in immunochemotherapy-treated diffuse large B-cell lymphoma
Abstract
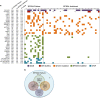
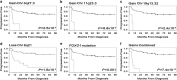
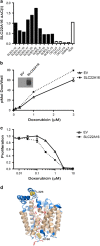
Lack of remission or early relapse remains a major clinical issue in diffuse large B-cell lymphoma (DLBCL), with 30% of patients failing standard of care. Although clinical factors and molecular signatures can partially predict DLBCL outcome, additional information is needed to identify high-risk patients, particularly biologic factors that might ultimately be amenable to intervention. Using whole-exome sequencing data from 51 newly diagnosed and immunochemotherapy-treated DLBCL patients, we evaluated the association of somatic genomic alterations with patient outcome, defined as failure to achieve event-free survival at 24 months after diagnosis (EFS24). We identified 16 genes with mutations, 374 with copy number gains and 151 with copy number losses that were associated with failure to achieve EFS24 (P<0.05). Except for FOXO1 and CIITA, known driver mutations did not correlate with EFS24. Gene losses were localized to 6q21-6q24.2, and gains to 3q13.12-3q29, 11q23.1-11q23.3 and 19q13.12-19q13.43. Globally, the number of gains was highly associated with poor outcome (P=7.4 × 10(-12)) and when combined with FOXO1 mutations identified 77% of cases that failed to achieve EFS24. One gene (SLC22A16) at 6q21, a doxorubicin transporter, was lost in 54% of EFS24 failures and our findings suggest it functions as a doxorubicin transporter in DLBCL cells.
Figures


Similar articles
-
Personalized risk prediction for event-free survival at 24 months in patients with diffuse large B-cell lymphoma.Am J Hematol. 2016 Feb;91(2):179-84. doi: 10.1002/ajh.24223. Epub 2015 Nov 26. Am J Hematol. 2016. PMID: 26492520 Free PMC article.
-
Mutations or copy number losses of CD58 and TP53 genes in diffuse large B cell lymphoma are independent unfavorable prognostic factors.Oncotarget. 2016 Dec 13;7(50):83294-83307. doi: 10.18632/oncotarget.13065. Oncotarget. 2016. PMID: 27825110 Free PMC article.
-
Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy.J Clin Oncol. 2014 Apr 1;32(10):1066-73. doi: 10.1200/JCO.2013.51.5866. Epub 2014 Feb 18. J Clin Oncol. 2014. PMID: 24550425 Free PMC article. Clinical Trial.
-
[Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese.
-
Next generation sequencing and the management of diffuse large B-cell lymphoma: from whole exome analysis to targeted therapy.Discov Med. 2014 Jul-Aug;18(97):51-65. Discov Med. 2014. PMID: 25091488 Review.
Cited by
-
SPIB is a novel prognostic factor in diffuse large B-cell lymphoma that mediates apoptosis via the PI3K-AKT pathway.Cancer Sci. 2016 Sep;107(9):1270-80. doi: 10.1111/cas.13001. Epub 2016 Sep 6. Cancer Sci. 2016. PMID: 27348272 Free PMC article.
-
Expression and Functional Contribution of Different Organic Cation Transporters to the Cellular Uptake of Doxorubicin into Human Breast Cancer and Cardiac Tissue.Int J Mol Sci. 2021 Dec 27;23(1):255. doi: 10.3390/ijms23010255. Int J Mol Sci. 2021. PMID: 35008681 Free PMC article.
-
Genetic analysis of B-cell lymphomas associated with hemophagocytic lymphohistiocytosis.Blood Adv. 2016 Dec 14;1(3):205-207. doi: 10.1182/bloodadvances.2016002006. eCollection 2016 Dec 27. Blood Adv. 2016. PMID: 29296936 Free PMC article. No abstract available.
-
Clinical Significance of Carnitine in the Treatment of Cancer: From Traffic to the Regulation.Oxid Med Cell Longev. 2023 Aug 10;2023:9328344. doi: 10.1155/2023/9328344. eCollection 2023. Oxid Med Cell Longev. 2023. PMID: 37600065 Free PMC article. Review.
-
Recent advances in CD5+ diffuse large B-cell lymphoma.Ann Hematol. 2024 Nov;103(11):4401-4412. doi: 10.1007/s00277-024-05974-8. Epub 2024 Aug 28. Ann Hematol. 2024. PMID: 39196380 Review.
References
-
- Howlader NNA, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, et al. (eds). SEER Cancer Statistics Review, 1975–2010 National Cancer Institute: Bethesda, MD; Available from: : http://seer.cancer.gov/csr/1975_2010/ (last accessed November 2012). SEER data submission, posted to the SEER website, April2013
-
- Sant M, Allemani C, Tereanu C, De Angelis R, Capocaccia R, Visser O, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116:3724–3734. - PubMed
-
- A predictive model for aggressive non-Hodgkin's lymphoma The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–994. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

