DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases
- PMID: 26315438
- PMCID: PMC4580133
- DOI: 10.1126/science.aab2666
DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases
Erratum in
-
Erratum for the Report "Base triplet stepping by the Rad51/RecA family of recombinases" by J. Y. Lee, T. Terakawa, Z. Qi, J. B. Steinfeld, S. Redding, Y. Kwon, W. A. Gaines, W. Zhao, P. Sung, E. C. Greene.Science. 2015 Oct 30;350(6260):aad6940. doi: 10.1126/science.aad6940. Science. 2015. PMID: 26516287 No abstract available.
Abstract
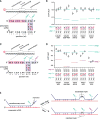
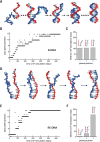
DNA strand exchange plays a central role in genetic recombination across all kingdoms of life, but the physical basis for these reactions remains poorly defined. Using single-molecule imaging, we found that bacterial RecA and eukaryotic Rad51 and Dmc1 all stabilize strand exchange intermediates in precise three-nucleotide steps. Each step coincides with an energetic signature (0.3 kBT) that is conserved from bacteria to humans. Triplet recognition is strictly dependent on correct Watson-Crick pairing. Rad51, RecA, and Dmc1 can all step over mismatches, but only Dmc1 can stabilize mismatched triplets. This finding provides insight into why eukaryotes have evolved a meiosis-specific recombinase. We propose that canonical Watson-Crick base triplets serve as the fundamental unit of pairing interactions during DNA recombination.
Copyright © 2015, American Association for the Advancement of Science.
Figures

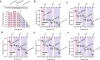
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

