Functional Restoration of gp91phox-Oxidase Activity by BAC Transgenesis and Gene Targeting in X-linked Chronic Granulomatous Disease iPSCs
- PMID: 26316390
- PMCID: PMC4886927
- DOI: 10.1038/mt.2015.154
Functional Restoration of gp91phox-Oxidase Activity by BAC Transgenesis and Gene Targeting in X-linked Chronic Granulomatous Disease iPSCs
Abstract
Chronic granulomatous disease (CGD) is an inherited immunodeficiency, caused by the inability of neutrophils to produce functional NADPH oxidase required for fighting microbial infections. The X-linked form of CGD (X-CGD), which is due to mutations in the CYBB (gp91phox) gene, a component of NADPH oxidase, accounts for about two-thirds of CGD cases. We derived induced pluripotent stem cells (iPSCs) from X-CGD patient keratinocytes using a Flp recombinase excisable lentiviral reprogramming vector. For restoring gp91phox function, we applied two strategies: transposon-mediated bacterial artificial chromosome (BAC) transgenesis and gene targeting using vectors with a fixed 5' homology arm (HA) of 8 kb and 3'HA varying in size from 30 to 80 kb. High efficiency of homologous recombination (up to 22%) was observed with increased size of the 3'HA. Both, BAC transgenesis and gene targeting resulted in functional restoration of the gp91phox measured by an oxidase activity assay in X-CGD iPSCs differentiated into the myeloid lineage. In conclusion, we delivered an important milestone towards the use of genetically corrected autologous cells for the treatment of X-CGD and monogenic diseases in general.
Figures

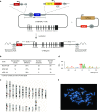


Similar articles
-
Lentivirus-mediated gene transfer of gp91phox corrects chronic granulomatous disease (CGD) phenotype in human X-CGD cells.J Gene Med. 2000 Sep-Oct;2(5):317-25. doi: 10.1002/1521-2254(200009/10)2:5<317::AID-JGM127>3.0.CO;2-P. J Gene Med. 2000. PMID: 11045425
-
TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells.Biomaterials. 2015 Nov;69:191-200. doi: 10.1016/j.biomaterials.2015.07.057. Epub 2015 Aug 3. Biomaterials. 2015. PMID: 26295532
-
Adenovirus-mediated gene transfer into monocyte-derived macrophages of patients with X-linked chronic granulomatous disease: ex vivo correction of deficient respiratory burst.Gene Ther. 1997 Jun;4(6):524-32. doi: 10.1038/sj.gt.3300432. Gene Ther. 1997. PMID: 9231068
-
[The X+ chronic granulomatous disease as a fabulous model to study the NADPH oxidase complex activation].Med Sci (Paris). 2007 May;23(5):526-32. doi: 10.1051/medsci/2007235526. Med Sci (Paris). 2007. PMID: 17502070 Review. French.
-
Progress in gene therapy for chronic granulomatous disease.J Infect Dis. 1999 Mar;179 Suppl 2:S318-25. doi: 10.1086/513852. J Infect Dis. 1999. PMID: 10081502 Review.
Cited by
-
Modeling blood diseases with human induced pluripotent stem cells.Dis Model Mech. 2019 Jun 4;12(6):dmm039321. doi: 10.1242/dmm.039321. Dis Model Mech. 2019. PMID: 31171568 Free PMC article. Review.
-
Stem Cell-Based Disease Models for Inborn Errors of Immunity.Cells. 2021 Dec 30;11(1):108. doi: 10.3390/cells11010108. Cells. 2021. PMID: 35011669 Free PMC article. Review.
-
Targeted Repair of CYBB in X-CGD iPSCs Requires Retention of Intronic Sequences for Expression and Functional Correction.Mol Ther. 2017 Feb 1;25(2):321-330. doi: 10.1016/j.ymthe.2016.11.012. Mol Ther. 2017. PMID: 28153086 Free PMC article.
-
Hematopoietic Cells from Pluripotent Stem Cells: Hope and Promise for the Treatment of Inherited Blood Disorders.Cells. 2022 Feb 5;11(3):557. doi: 10.3390/cells11030557. Cells. 2022. PMID: 35159366 Free PMC article. Review.
-
Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors.Genes (Basel). 2020 May 6;11(5):511. doi: 10.3390/genes11050511. Genes (Basel). 2020. PMID: 32384610 Free PMC article.
References
-
- Winkelstein, JA, Marino, MC, Johnston, RB Jr., Boyle, J, Curnutte, J, Gallin, JI et al. (2000). Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79: 155–169. - PubMed
-
- Noack, D, Heyworth, PG, Newburger, PE and Cross, AR (2001). An unusual intronic mutation in the CYBB gene giving rise to chronic granulomatous disease. Biochim Biophys Acta 1537: 125–131. - PubMed
-
- Heyworth, PG, Curnutte, JT, Rae, J, Noack, D, Roos, D, van Koppen, E et al. (2001). Hematologically important mutations: X-linked chronic granulomatous disease (second update). Blood Cells Mol Dis 27: 16–26. - PubMed
-
- Noack, D, Heyworth, PG, Curnutte, JT, Rae, J and Cross, AR (1999). A novel mutation in the CYBB gene resulting in an unexpected pattern of exon skipping and chronic granulomatous disease. Biochim Biophys Acta 1454: 270–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

