Selectivity in the Use of Gi/o Proteins Is Determined by the DRF Motif in CXCR6 and Is Cell-Type Specific
- PMID: 26316539
- PMCID: PMC4613941
- DOI: 10.1124/mol.115.099960
Selectivity in the Use of Gi/o Proteins Is Determined by the DRF Motif in CXCR6 and Is Cell-Type Specific
Abstract
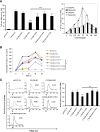
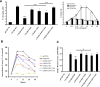
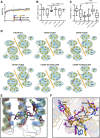
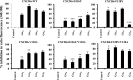
CXCR6, the receptor for CXCL16, is expressed on multiple cell types and can be a coreceptor for human immunodeficiency virus 1. Except for CXCR6, all human chemokine receptors contain the D(3.49)R(3.50)Y(3.51) sequence, and all but two contain A(3.53) at the cytoplasmic terminus of the third transmembrane helix (H3C), a region within class A G protein-coupled receptors that contacts G proteins. In CXCR6, H3C contains D(3.49)R(3.50)F(3.51)I(3.52)V(3.53) at positions 126-130. We investigated the importance and interdependence of the canonical D126 and the noncanonical F128 and V130 in CXCR6 by mutating D126 to Y, F128 to Y, and V130 to A singly and in combination. For comparison, we mutated the analogous positions D142, Y144, and A146 to Y, F, and V, respectively, in CCR6, a related receptor containing the canonical sequences. Mutants were analyzed in both human embryonic kidney 293T and Jurkat E6-1 cells. Our data show that for CXCR6 and/or CCR6, mutations in H3C can affect both receptor signaling and chemokine binding; noncanonical H3C sequences are functionally linked, with dual changes mitigating the effects of single mutations; mutations in H3C that compromise receptor activity show selective defects in the use of individual Gi/o proteins; and the effects of mutations in H3C on receptor function and selectivity in Gi/o protein use can be cell-type specific. Our findings indicate that the ability of CXCR6 to make promiscuous use of the available Gi/o proteins is exquisitely dependent on sequences within the H3C and suggest that the native sequence allows for preservation of this function across different cellular environments.
U.S. Government work not protected by U.S. copyright.
Figures



References
-
- Acharya S, Saad Y, Karnik SS. (1997) Transducin-alpha C-terminal peptide binding site consists of C-D and E-F loops of rhodopsin. J Biol Chem 272:6519–6524. - PubMed
-
- Alkhatib G, Liao F, Berger EA, Farber JM, Peden KW. (1997) A new SIV co-receptor, STRL33. Nature 388:238. - PubMed
-
- Arai H, Charo IF. (1996) Differential regulation of G-protein-mediated signaling by chemokine receptors. J Biol Chem 271:21814–21819. - PubMed
-
- Auger GA, Pease JE, Shen X, Xanthou G, Barker MD. (2002) Alanine scanning mutagenesis of CCR3 reveals that the three intracellular loops are essential for functional receptor expression. Eur J Immunol 32:1052–1058. - PubMed
-
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, et al. (2013) International Union of Pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev 66:1–79. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

