The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses
- PMID: 26316841
- PMCID: PMC4544635
- DOI: 10.1007/s11032-014-0165-5
The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses
Abstract
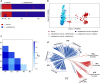
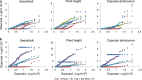
Pale flax (Linum bienne Mill.) is the wild progenitor of cultivated flax (Linum usitatissimum L.) and represents the primary gene pool to broaden its genetic base. Here, a collection of 125 pale flax accessions and the Canadian flax core collection of 407 accessions were genotyped using 112 genome-wide simple sequence repeat markers and phenotyped for nine traits with the aim of conducting population structure, molecular diversity and association mapping analyses. The combined population structure analysis identified two well-supported major groups corresponding to pale and cultivated flax. The L. usitatissimum convar. crepitans accessions most closely resembled its wild progenitor, both having dehiscent capsules. The unbiased Nei's genetic distance (0.65) confirmed the strong genetic differentiation between cultivated and pale flax. Similar levels of genetic diversity were observed in both species, albeit 430 (48 %) of pale flax alleles were unique, in agreement with their high genetic differentiation. Significant associations were identified for seven and four traits in pale and cultivated flax, respectively. Favorable alleles with potentially positive effect to improve yield through yield components were identified in pale flax. The allelic frequencies of markers associated with domestication-related traits such as capsular dehiscence indicated directional selection with the most common alleles in pale flax being absent or rare in cultivated flax and vice versa. Our results demonstrated that pale flax is a potential source of novel variation to improve multiple traits in cultivated flax and that association mapping is a suitable approach to screening pale flax germplasm to identify favorable quantitative trait locus alleles.
Keywords: Association mapping; Cultivated flax; Exotic alleles; Genetic diversity; Pale flax; Wild ancestor.
Figures

References
-
- Allaby RG, Peterson GW, Merriwether DA, Fu Y-B. Evidence of the domestication history of flax (Linum usitatissimum) from genetic diversity of the sad2 locus. Theor Appl Genet. 2005;112:58–65. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
-
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. - PubMed
-
- Caballero A, Toro MA. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Genet. 2002;3:289–299.
LinkOut - more resources
Full Text Sources
Other Literature Sources

